3DEO
 
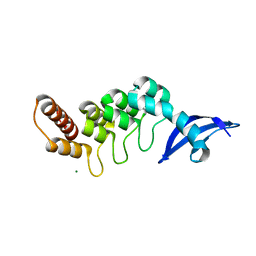 | |
3B9Q
 
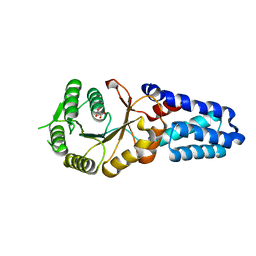 | | The crystal structure of cpFtsY from Arabidopsis thaliana | | Descriptor: | Chloroplast SRP receptor homolog, alpha subunit CPFTSY, MALONATE ION | | Authors: | Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2007-11-06 | | Release date: | 2007-12-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of the chloroplast signal recognition particle (SRP) receptor reveals mechanistic details of SRP GTPase activation and a conserved membrane targeting site
Febs Lett., 581, 2007
|
|
4LRI
 
 | | Anti CMV Fab Fragment | | Descriptor: | MSL-109 Heavy Chain, MSL-109 Light Chain | | Authors: | Stengel, K.F. | | Deposit date: | 2013-07-19 | | Release date: | 2014-05-21 | | Last modified: | 2014-07-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism for neutralizing activity by the anti-CMV gH/gL monoclonal antibody MSL-109.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3GT8
 
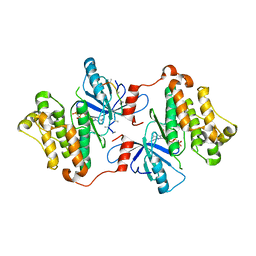 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
3ROF
 
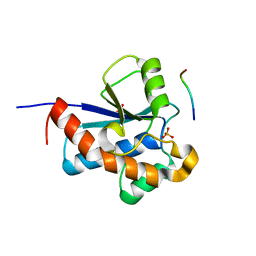 | | Crystal Structure of the S. aureus Protein Tyrosine Phosphatase PtpA | | Descriptor: | Expression tag cleaved from protein-tyrosine-phosphatase ptpA, Low molecular weight protein-tyrosine-phosphatase ptpA, PHOSPHATE ION | | Authors: | Grundner, C, Chou, S, Engel, K. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure and substrate recognition of the Staphylococcus aureus protein tyrosine phosphatase PtpA.
J.Mol.Biol., 413, 2011
|
|
3UCR
 
 | | Crystal structure of the immunoreceptor TIGIT IgV domain | | Descriptor: | CHLORIDE ION, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Yin, J.P, Stengel, K.F, Rouge, L, Bazan, J.F, Wiesmann, C. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-14 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.627 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UDW
 
 | | Crystal structure of the immunoreceptor TIGIT in complex with Poliovirus receptor (PVR/CD155/necl-5) D1 domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Poliovirus receptor, T cell immunoreceptor with Ig and ITIM domains | | Authors: | Rouge, L, Stengel, K.F, Yin, J.P, Bazan, F.J, Wiesmann, C. | | Deposit date: | 2011-10-28 | | Release date: | 2012-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure of TIGIT immunoreceptor bound to poliovirus receptor reveals a cell-cell adhesion and signaling mechanism that requires cis-trans receptor clustering.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3DEP
 
 | | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43 | | Descriptor: | CHLORIDE ION, Signal recognition particle 43 kDa protein, YPGGSFDPLGLA | | Authors: | Holdermann, I, Stengel, K.F, Wild, K, Sinning, I. | | Deposit date: | 2008-06-10 | | Release date: | 2008-08-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for specific substrate recognition by the chloroplast signal recognition particle protein cpSRP43.
Science, 321, 2008
|
|
