6V0H
 
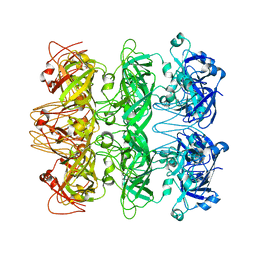 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 6 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6XBD
 
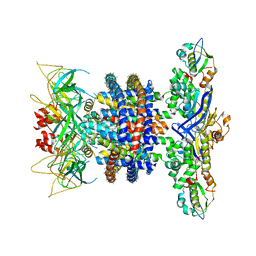 | | Cryo-EM structure of MlaFEDB in nanodiscs with phospholipid substrates | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, MSP1D1, Phospholipid ABC transporter permease protein MlaE, ... | | Authors: | Coudray, N, Isom, G.L, MacRae, M.R, Saiduddin, M, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2020-06-05 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structure of bacterial phospholipid transporter MlaFEDB with substrate bound.
Elife, 9, 2020
|
|
6V0C
 
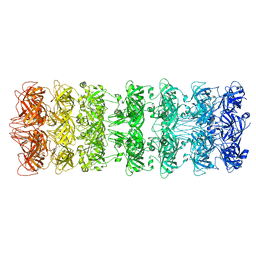 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 1 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0E
 
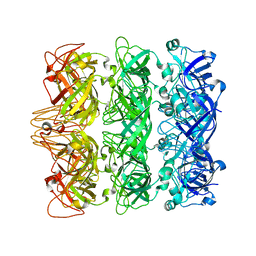 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 3 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0J
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 8 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0G
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 5 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0I
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 7 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6VCI
 
 | | Lipophilic envelope-spanning tunnel protein (LetB), domains MCE2-MCE3 | | Descriptor: | Lipophilic envelope-spanning tunnel protein LetB | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-12-21 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V0D
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 2 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
7UDK
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 bound to LRPx4 peptide | | Descriptor: | 4xLRP, Designed helical repeat protein (DHR) RPB_LRP2_R4 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDN
 
 | |
7UDM
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 in alternative conformation 1 (with peptide) | | Descriptor: | 6xPLP, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
3QL3
 
 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
7UDL
 
 | | Crystal structure of designed helical repeat protein RPB_PLP1_R6 bound to PLPx6 peptide | | Descriptor: | 1,2-ETHANEDIOL, 6xPLP Peptide, Designed helical repeat protein (DHR) RPB_PLP1_R6 | | Authors: | Chang, Y, Redler, R.L, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UDO
 
 | | Crystal structure of designed helical repeat protein RPB_LRP2_R4 (proteolysis fragment?), forming pseudopolymeric filaments | | Descriptor: | 1,2-ETHANEDIOL, Designed helical repeat protein (DHR) RPB_LRP2_R4, PHOSPHATE ION | | Authors: | Redler, R.L, Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2022-03-20 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of modular peptide-binding proteins by superhelical matching.
Nature, 616, 2023
|
|
7UE2
 
 | |
4M6K
 
 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and folate | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, GLYCEROL, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.396 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3QL0
 
 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
6WYR
 
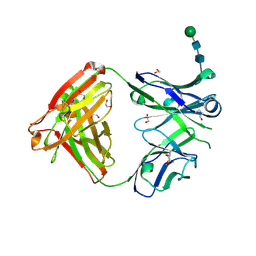 | | Crystal structure of anti-Muscle Specific Kinase (MuSK) Fab, MuSK1A | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MuSK1A heavy chain, ... | | Authors: | Vieni, C, Ekiert, D. | | Deposit date: | 2020-05-13 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Affinity maturation is required for pathogenic monovalent IgG4 autoantibody development in myasthenia gravis.
J.Exp.Med., 217, 2020
|
|
6XSS
 
 | | CryoEM structure of designed helical fusion protein C4_nat_HFuse-7900 | | Descriptor: | C4_nat_HFuse-7900 | | Authors: | Redler, R.L, Edman, N.I, Baker, D, Ekiert, D, Bhabha, G. | | Deposit date: | 2020-07-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Design of multi-scale protein complexes by hierarchical building block fusion.
Nat Commun, 12, 2021
|
|
5UW8
 
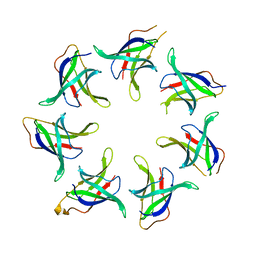 | |
4M6L
 
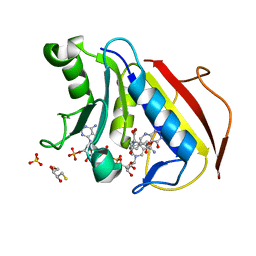 | | Crystal structure of human dihydrofolate reductase (DHFR) bound to NADP+ and 5,10-dideazatetrahydrofolic acid | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Dihydrofolate reductase, N-(4-{2-[(6S)-2-amino-4-oxo-1,4,5,6,7,8-hexahydropyrido[2,3-d]pyrimidin-6-yl]ethyl}benzoyl)-L-glutamic acid, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2013-08-09 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Divergent evolution of protein conformational dynamics in dihydrofolate reductase.
Nat.Struct.Mol.Biol., 20, 2013
|
|
6XGZ
 
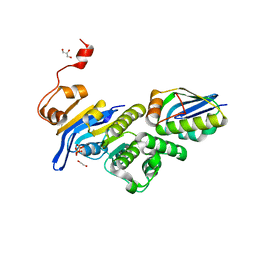 | | Crystal structure of E. coli MlaFB ABC transport subunits in the monomeric state | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter maintaining OM lipid asymmetry, cytoplasmic STAS component, ... | | Authors: | Chang, Y, Bhabha, G, Ekiert, D.C. | | Deposit date: | 2020-06-18 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of MlaFB uncovers novel mechanisms of ABC transporter regulation.
Elife, 9, 2020
|
|
5J8E
 
 | |
5CWC
 
 | |
