7ZEM
 
 | |
7ZEO
 
 | |
7ZEK
 
 | |
7SZI
 
 | | Cryo-EM structure of OmpK36-TraN mating pair stabilization proteins from carbapenem-resistant Klebsiella pneumoniae | | Descriptor: | OmpK36, TraN | | Authors: | Beltran, L.C, Seddon, C, Beis, K, Frankel, G, Egelman, E.H. | | Deposit date: | 2021-11-27 | | Release date: | 2022-06-08 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Mating pair stabilization mediates bacterial conjugation species specificity.
Nat Microbiol, 7, 2022
|
|
8ABD
 
 | |
8ABN
 
 | |
6ZL2
 
 | |
8A7Z
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid polyketide synthase-non ribosomal peptide synthetase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8AIG
 
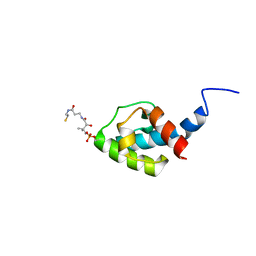 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-07-26 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8ALL
 
 | | NMR structure of holo-acp | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Hybrid non ribosomal peptide synthetase-polyketide synthase | | Authors: | Collin, S, Weissman, K.J, Chagot, B, Gruez, A. | | Deposit date: | 2022-08-01 | | Release date: | 2023-03-22 | | Last modified: | 2023-03-29 | | Method: | SOLUTION NMR | | Cite: | Decrypting the programming of beta-methylation in virginiamycin M biosynthesis.
Nat Commun, 14, 2023
|
|
8S1W
 
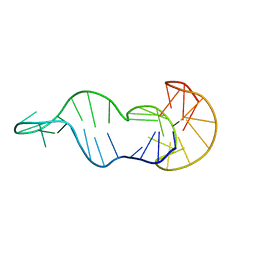 | |
3HJH
 
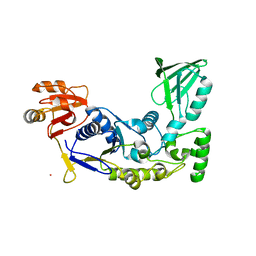 | | A rigid N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor | | Descriptor: | COBALT (II) ION, Transcription-repair-coupling factor | | Authors: | Murphy, M, Gong, P, Ralto, K, Manelyte, L, Savery, N, Theis, K. | | Deposit date: | 2009-05-21 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An N-terminal clamp restrains the motor domains of the bacterial transcription-repair coupling factor Mfd.
Nucleic Acids Res., 37, 2009
|
|
7ATZ
 
 | |
8PSE
 
 | |
2XQZ
 
 | | Neutron structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | BETA-LACTAMSE TOHO-1 | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-05-08 | | Method: | NEUTRON DIFFRACTION (2.1 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
5KMW
 
 | | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex | | Descriptor: | Beta-lactamase Toho-1, OPEN FORM - PENICILLIN G, PENICILLIN G, ... | | Authors: | Coates, L, Langan, P.S, Vandavasi, V.G, Weiss, K.L, Cooper, J.B, Ginell, S.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-03-01 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | TOHO1 Beta lactamase mutant E166A/R274N/R276N -benzyl penicillin complex
to be published
|
|
5G18
 
 | | Direct Observation of Active-site Protonation States in a Class A beta lactamase with a monobactam substrate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BETA-LACTAMASE CTX-M-97, SULFATE ION | | Authors: | Vandavasi, V.G, Weiss, K.L, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
8R4W
 
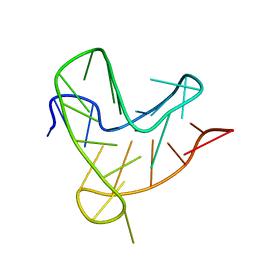 | |
6ZTE
 
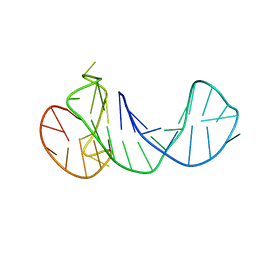 | |
8RAJ
 
 | | NMR structure of PKS docking domains | | Descriptor: | Beta-ketoacyl synthase, Trimethylamine monooxygenase | | Authors: | Scat, S, Weissman, K.J, Chagot, B. | | Deposit date: | 2023-12-01 | | Release date: | 2024-06-05 | | Last modified: | 2024-07-17 | | Method: | SOLUTION NMR | | Cite: | Insights into docking in megasynthases from the investigation of the toblerol trans -AT polyketide synthase: many alpha-helical means to an end.
Rsc Chem Biol, 5, 2024
|
|
2XR0
 
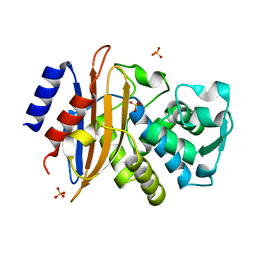 | | Room temperature X-ray structure of the perdeuterated Toho-1 R274N R276N double mutant beta-lactamase | | Descriptor: | SULFATE ION, TOHO-1 BETA-LACTAMASE | | Authors: | Tomanicek, S.J, Wang, K.K, Weiss, K.L, Blakeley, M.P, Cooper, J, Chen, Y, Coates, L. | | Deposit date: | 2010-09-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Active Site Protonation States of Perdeuterated Toho-1 Beta-Lactamase Determined by Neutron Diffraction Support a Role for Glu166 as the General Base in Acylation.
FEBS Lett., 585, 2011
|
|
1AUK
 
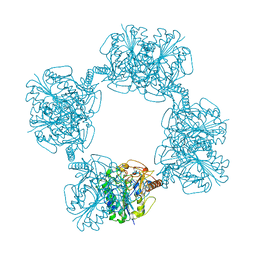 | | HUMAN ARYLSULFATASE A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ARYLSULFATASE A, MAGNESIUM ION | | Authors: | Lukatela, G, Krauss, N, Theis, K, Gieselmann, V, Von Figura, K, Saenger, W. | | Deposit date: | 1997-08-29 | | Release date: | 1998-03-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human arylsulfatase A: the aldehyde function and the metal ion at the active site suggest a novel mechanism for sulfate ester hydrolysis.
Biochemistry, 37, 1998
|
|
6F7L
 
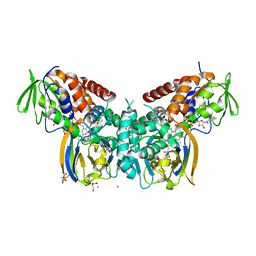 | | Crystal structure of LkcE R326Q mutant in complex with its substrate | | Descriptor: | ACETATE ION, Amine oxidase LkcE, CALCIUM ION, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-12-11 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
6F32
 
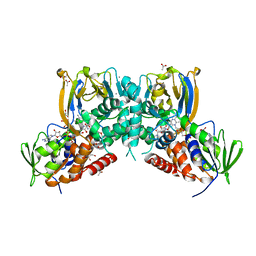 | | Crystal structure of a dual function amine oxidase/cyclase in complex with substrate analogues | | Descriptor: | (2~{R},3~{R})-2,3-bis(oxidanyl)-~{N},~{N}'-dipropyl-butanediamide, ACETATE ION, Amine oxidase LkcE, ... | | Authors: | Dorival, J, Risser, F, Jacob, C, Collin, S, Drager, G, Kirschning, A, Paris, C, Chagot, B, Gruez, A, Weissman, K.J. | | Deposit date: | 2017-11-27 | | Release date: | 2018-09-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insights into a dual function amide oxidase/macrocyclase from lankacidin biosynthesis.
Nat Commun, 9, 2018
|
|
1ORM
 
 | |
