5HSS
 
 | | Linalool dehydratase/isomerase: Ldi with monoterpene substrate | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Beta-Myrcene, Geraniol, ... | | Authors: | Weidenweber, S, Marmulla, R, Harder, J, Ermler, U. | | Deposit date: | 2016-01-26 | | Release date: | 2016-04-27 | | Last modified: | 2016-05-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis.
Febs Lett., 590, 2016
|
|
5HLR
 
 | | Linalool dehydratase/isomerase: Ldi-apo | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Linalool dehydratase/isomerase | | Authors: | Weidenweber, S, Marmulla, R, Harder, J, Ermler, U. | | Deposit date: | 2016-01-15 | | Release date: | 2016-04-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.911 Å) | | Cite: | X-ray structure of linalool dehydratase/isomerase from Castellaniella defragrans reveals enzymatic alkene synthesis.
Febs Lett., 590, 2016
|
|
7AYX
 
 | |
3KOF
 
 | | Crystal structure of the double mutant F178Y/R181E of E.coli transaldolase B | | Descriptor: | SULFATE ION, Transaldolase B | | Authors: | Schneider, S, Gutierrez, M, Sandalova, T, Schneider, G, Clapes, P, Sprenger, G.A, Samland, A.K. | | Deposit date: | 2009-11-13 | | Release date: | 2010-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning the Active Site of Transaldolase TalB from Escherichia coli: New Variants with Improved Affinity towards Nonphosphorylated Substrates.
Chembiochem, 11, 2010
|
|
8FAI
 
 | | Cryo-EM structure of the Agrobacterium T-pilus | | Descriptor: | (7Z,19R,22S,25R)-22,25,26-trihydroxy-16,22-dioxo-17,21,23-trioxa-22lambda~5~-phosphahexacos-7-en-19-yl (9Z)-octadec-9-enoate, Protein virB2 | | Authors: | Kreida, S, Narita, A, Johnson, M.D, Tocheva, E.I, Das, A, Jensen, G.J, Ghosal, D. | | Deposit date: | 2022-11-27 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structure of the Agrobacterium tumefaciens T4SS-associated T-pilus reveals stoichiometric protein-phospholipid assembly.
Structure, 31, 2023
|
|
7Z06
 
 | |
6S8Z
 
 | |
4GI2
 
 | | Crotonyl-CoA Carboxylase/Reductase | | Descriptor: | Crotonyl-CoA carboxylase/reductase, MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Weidenweber, S, Erb, T.J, Ermler, U. | | Deposit date: | 2012-08-08 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crotonyl-CoA Carboxylase/Reductase
To be Published
|
|
6YX5
 
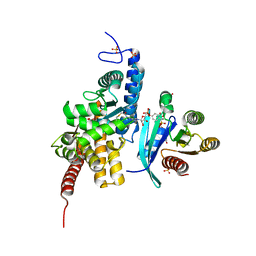 | | Structure of DrrA from Legionella pneumophilia in complex with human Rab8a | | Descriptor: | MAGNESIUM ION, Multifunctional virulence effector protein DrrA, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Schneider, S, Du, J, von Wrisberg, M.K, Lang, K, Itzen, A. | | Deposit date: | 2020-04-30 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Rab1-AMPylation by Legionella DrrA is allosterically activated by Rab1.
Nat Commun, 12, 2021
|
|
8R33
 
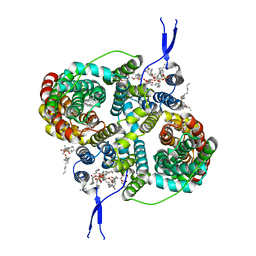 | |
8R35
 
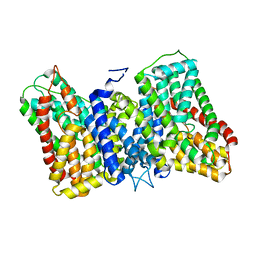 | |
8R34
 
 | | CryoEM structure of the symmetric Pho90 dimer from yeast with substrates. | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, Low-affinity phosphate transporter PHO90, PHOSPHATE ION, ... | | Authors: | Schneider, S, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2023-11-08 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Complementary structures of the yeast phosphate transporter Pho90 provide insights into its transport mechanism.
Structure, 32, 2024
|
|
8Q91
 
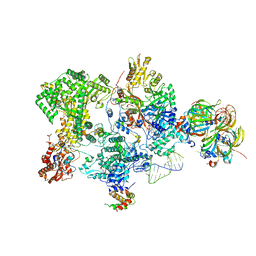 | | Structure of the human 20S U5 snRNP core | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-08-19 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8RC0
 
 | | Structure of the human 20S U5 snRNP | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CD2 antigen cytoplasmic tail-binding protein 2, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Schneider, S, Galej, W.P. | | Deposit date: | 2023-12-05 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human 20S U5 snRNP.
Nat.Struct.Mol.Biol., 31, 2024
|
|
1EM0
 
 | | COMPLEX OF D(CCTAGG) WITH TETRA-[N-METHYL-PYRIDYL] PORPHYRIN | | Descriptor: | DNA (5'-D(*(CBR)P*CP*TP*AP*GP*G)-3'), MAGNESIUM ION, TETRA[N-METHYL-PYRIDYL] PORPHYRIN-NICKEL | | Authors: | Neidle, S, Sanderson, M, Bennett, M, Krah, A, Wien, F, Garman, E, McKenna, R. | | Deposit date: | 2000-03-14 | | Release date: | 2000-08-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.9 Å) | | Cite: | A DNA-porphyrin minor-groove complex at atomic resolution: the structural consequences of porphyrin ruffling.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5JM0
 
 | | Structure of the S. cerevisiae alpha-mannosidase 1 | | Descriptor: | Alpha-mannosidase,Alpha-mannosidase,Alpha-mannosidase | | Authors: | Schneider, S, Kosinski, J, Jakobi, A.J, Hagen, W.J.H, Sachse, C. | | Deposit date: | 2016-04-28 | | Release date: | 2016-06-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle.
Embo Rep., 17, 2016
|
|
2V79
 
 | | Crystal Structure of the N-terminal domain of DnaD from Bacillus Subtilis | | Descriptor: | CHLORIDE ION, DNA REPLICATION PROTEIN DNAD, SODIUM ION | | Authors: | Schneider, S, Zhang, W, Soultanas, P, Paoli, M. | | Deposit date: | 2007-07-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the N-Terminal Oligomerization Domain of Dnad Reveals a Unique Tetramerization Motif and Provides Insights Into Scaffold Formation.
J.Mol.Biol., 376, 2008
|
|
6QRI
 
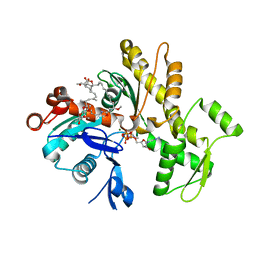 | | Structure of rabbit G-actin in complex with chivosazole A | | Descriptor: | (2~{R},3~{R},5~{S},6~{E},8~{E},10~{Z},12~{S},13~{R},16~{Z},18~{E},20~{Z},22~{E},24~{R},25~{S},26~{E},28~{Z})-13-[(2~{S},3~{S},5~{S})-3,5-bis(oxidanyl)hexan-2-yl]-25-[(2~{R},3~{R},4~{S},5~{R},6~{R})-3,4-dimethoxy-6-methyl-5-oxidanyl-oxan-2-yl]oxy-3-methoxy-2,12,22,24-tetramethyl-5-oxidanyl-14,32-dioxa-33-azabicyclo[28.2.1]tritriaconta-1(33),6,8,10,16,18,20,22,26,28,30-undecaen-15-one, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Schneider, S, Wang, S, Zahler, S. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chivosazole A Modulates Protein-Protein Interactions of Actin.
J.Nat.Prod., 82, 2019
|
|
5G5S
 
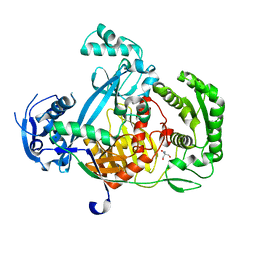 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ARGONAUTE, MAGNESIUM ION | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
5LBT
 
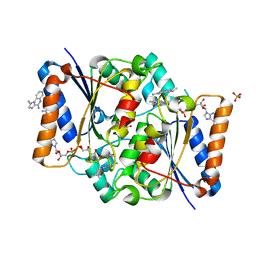 | | Structure of the human quinone reductase 2 (NQO2) in complex with imiquimod | | Descriptor: | 1-(2-methylpropyl)imidazo[4,5-c]quinolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Gross, O. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Imiquimod Inhibits Mitochondrial Complex I and Induces K+ efflux-independent Nlrp3 Inflammasome Activation via Nek7
To Be Published
|
|
5LBU
 
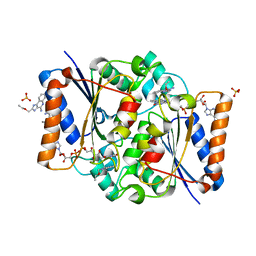 | | Structure of the human quinone reductase 2 (NQO2) in complex with to CL097 | | Descriptor: | 2-(ethoxymethyl)-1H-imidazo[4,5-c]quinolin-4-amine, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Schneider, S, Gross, O. | | Deposit date: | 2016-06-17 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Imiquimod Inhibits Mitochondrial Complex I and Induces K+ efflux-independent Nlrp3 Inflammasome Activation via Nek7
To Be Published
|
|
5LCL
 
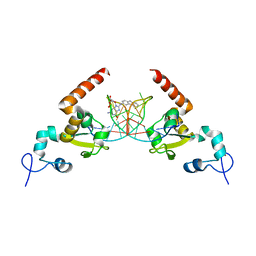 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH C8-aminofluorene- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, GCTCTAC(8AF)TCATCA, ... | | Authors: | Schneider, S, Carell, T, Ebert, C, Simon, N. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
5LCM
 
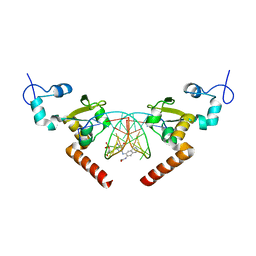 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH N2-acetylaminonaphtyl- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*CP*TP*AP*CP*(AAN)P*TP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, ... | | Authors: | Schneider, S, Ebert, C, Simon, N, Carell, T. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
5G5T
 
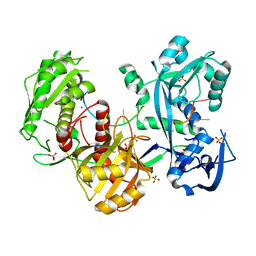 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii in complex with guide DNA | | Descriptor: | ARGONAUTE, GUIDE DNA, MAGNESIUM ION, ... | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
2KD3
 
 | |
