2ELX
 
 | | Solution structure of the 8th C2H2 zinc finger of mouse Zinc finger protein 406 | | Descriptor: | ZINC ION, Zinc finger protein 406 | | Authors: | Tochio, N, Yoneyama, M, Koshiba, S, Watanabe, S, Harada, T, Umehara, T, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-27 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the 8th C2H2 zinc finger of mouse Zinc finger protein 406
To be Published
|
|
2CXL
 
 | | RUN domain of Rap2 interacting protein x, crystallized in I422 space group | | Descriptor: | rap2 interacting protein x | | Authors: | Kukimoto-Niino, M, Umehara, T, Murayama, K, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-30 | | Release date: | 2005-12-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the RUN Domain of the RAP2-interacting Protein x
J.Biol.Chem., 281, 2006
|
|
2CY1
 
 | | Crystal structure of APE1850 | | Descriptor: | NusA protein homolog | | Authors: | Shibata, R, Bessho, Y, Umehara, T, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-07-04 | | Release date: | 2006-01-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallization of the archaeal transcription termination factor NusA: a significant decrease in twinning under microgravity conditions
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2COB
 
 | | Solution structures of the HTH domain of human LCoR protein | | Descriptor: | LCoR protein | | Authors: | Nameki, N, Umehara, T, Sato, M, Koshiba, S, Inoue, M, Tanaka, A, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the HTH domain of human LCoR protein
To be Published
|
|
2RS9
 
 | | Solution structure of the bromodomain of human BRPF1 in complex with histone H4K5ac peptide | | Descriptor: | Acetylated lysine 5 of peptide from Histone H4, Peregrin | | Authors: | Qin, X, Nagashima, T, Umehara, T, Hayashi, F, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-12 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Site-specific histone recognition by the bromodomain of Brpf1 and the role in MOZ/MORF histone acetyltransferase complexes
To be Published
|
|
5X18
 
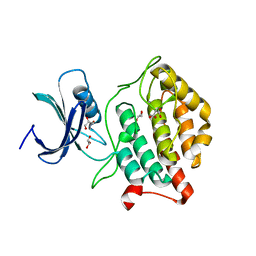 | | Crystal structure of Casein kinase I homolog 1 | | Descriptor: | Casein kinase I homolog 1, GLYCEROL, MALONIC ACID | | Authors: | Kikuchi, M, Shinohara, Y, Ueda, H.R, Umehara, T. | | Deposit date: | 2017-01-25 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock
Mol. Cell, 67, 2017
|
|
2EJR
 
 | | LSD1-tranylcypromine complex | | Descriptor: | Lysine-specific histone demethylase 1, [(2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL (2R,3S,4S)-5-[7,8-DIMETHYL-2,4-DIOXO-5-(3-PHENYLPROPANOYL)-1,3,4,5-TETRAHYDROBENZO[G]PTERIDIN-10(2H)-YL]-2,3,4-TRIHYDROXYPENTYL DIHYDROGEN DIPHOSPHATE | | Authors: | Sengoku, T, Mimasu, S, Umehara, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-20 | | Release date: | 2008-01-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25A
Biochem.Biophys.Res.Commun., 366, 2008
|
|
7EIC
 
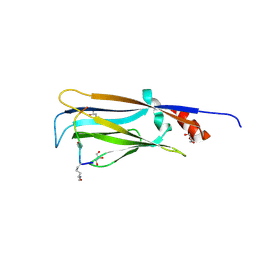 | |
7EID
 
 | |
7EIE
 
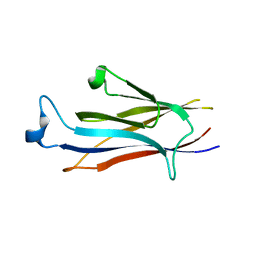 | | Crystal structure of YEATS2 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 2 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Elucidation of binding preferences of YEATS domains to site-specific acetylated nucleosome core particles.
J.Biol.Chem., 298, 2022
|
|
2DVS
 
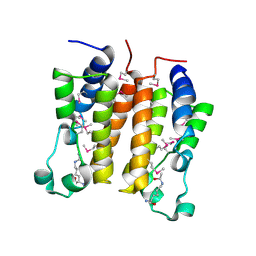 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVR
 
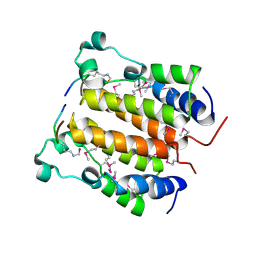 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
2DVQ
 
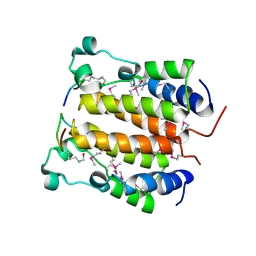 | | Crystal structure analysis of the N-terminal bromodomain of human BRD2 complexed with acetylated histone H4 peptide | | Descriptor: | Bromodomain-containing protein 2, histone H4 | | Authors: | Nakamura, Y, Umehara, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-08-01 | | Release date: | 2007-08-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Basis for Acetylated Histone H4 Recognition by the Human BRD2 Bromodomain.
J.Biol.Chem., 285, 2010
|
|
3WN2
 
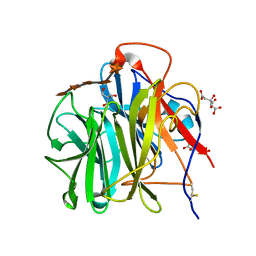 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylohexaose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WMY
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3W5M
 
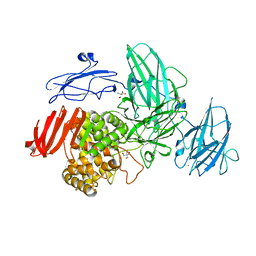 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, Putative rhamnosidase | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3WMZ
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase ethylmercury derivative | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ETHYL MERCURY ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3W5N
 
 | | Crystal Structure of Streptomyces avermitilis alpha-L-rhamnosidase complexed with L-rhamnose | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Putative rhamnosidase, ... | | Authors: | Fujimoto, Z, Jackson, A, Michikawa, M, Maehara, T, Momma, M, Henrissat, B.F, Gilbert, H.J, Kaneko, S. | | Deposit date: | 2013-01-31 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of a Streptomyces avermitilis alpha-L-rhamnosidase reveals a novel carbohydrate-binding module CBM67 within the six-domain arrangement.
J.Biol.Chem., 288, 2013
|
|
3WN1
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with xylotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
3WN0
 
 | | Crystal Structure of Streptomyces coelicolor alpha-L-arabinofuranosidase in complex with L-arabinose | | Descriptor: | CALCIUM ION, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Fujimoto, Z, Maehara, T, Ichinose, H, Michikawa, M, Harazono, K, Kaneko, S. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and characterization of the glycoside hydrolase family 62 alpha-L-arabinofuranosidase from Streptomyces coelicolor
J.Biol.Chem., 289, 2014
|
|
