2LSM
 
 | | Solution structure of gpFI C-terminal domain | | Descriptor: | DNA-packaging protein FI | | Authors: | Popovic, A, Wu, B, Edwards, A.M, Davidson, A.R, Maxwell, K.L. | | Deposit date: | 2012-05-02 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and biochemical characterization of phage lambda FI protein (gpFI) reveals a novel mechanism of DNA packaging chaperone activity.
J.Biol.Chem., 287, 2012
|
|
3L9A
 
 | | Structure of the C-terminal domain from a Streptococcus mutans hypothetical | | Descriptor: | GLYCEROL, SODIUM ION, uncharacterized protein | | Authors: | Singer, A.U, Cuff, M.E, Xu, X, Cui, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-04 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the C-terminal domain from a Streptococcus mutans hypothetical
To be Published
|
|
3LLT
 
 | | Crystal structure of PF14_0431, kinase domain. | | Descriptor: | ACETATE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Loppnau, P, MacKenzie, F, Sullivan, H, Weadge, J, Kozieradzki, I, Cossar, D, Sinesterra, G, Vedadi, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Hui, R, Qiu, W, Hutchinson, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-01-29 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of PF14_0431, kinase domain.
TO BE PUBLISHED
|
|
3LHH
 
 | | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1. | | Descriptor: | ADENOSINE MONOPHOSPHATE, CBS domain protein | | Authors: | Tan, K, Kagan, O, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-02-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of CBS domain protein from Shewanella oneidensis MR-1.
To be Published
|
|
3LEQ
 
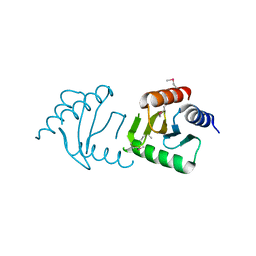 | | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A | | Descriptor: | uncharacterized protein cvnB5 | | Authors: | Stein, A.J, Xu, X, Cui, H, Ng, J, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-01-15 | | Release date: | 2010-02-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Crystal Structure of the Roadblock/LC7 domain from Streptomyces avermitillis to 1.85A
To be Published
|
|
6S4L
 
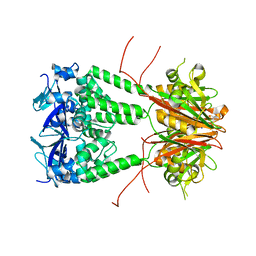 | | Structure of human KCTD1 | | Descriptor: | BTB/POZ domain-containing protein KCTD1, IODIDE ION, SODIUM ION | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Pike, A.C.W, Newman, J.A, Krojer, T, Shrestha, L, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C, Edwards, A, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure of human KCTD1
To be published
|
|
6V2D
 
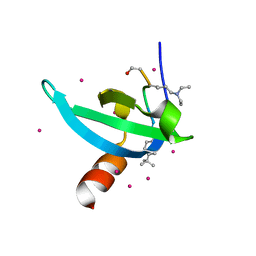 | | Crystal Structure of chromodomain of CDYL2 in complex with inhibitor UNC3866 | | Descriptor: | Chromodomain Y-like protein 2, UNC3866, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-22 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
3LU2
 
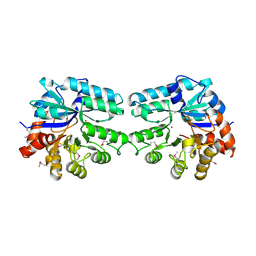 | | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase | | Descriptor: | Lmo2462 protein, ZINC ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of lmo2462, a Listeria monocytogenes amidohydrolase family putative dipeptidase
To be Published
|
|
6V41
 
 | | crystal structure of CDY1 chromodomain bound to H3K9me3 | | Descriptor: | Histone H3.1 Peptide, Testis-specific chromodomain protein Y 1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Walker, J.R, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-11-27 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.603 Å) | | Cite: | Structural Basis for the Binding Selectivity of Human CDY Chromodomains.
Cell Chem Biol, 27, 2020
|
|
6D1T
 
 | | Complex of MBD1-MBD and methylated DNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*AP*(5CM)P*GP*TP*TP*GP*GP*C)-3'), Methyl-CpG-binding domain protein 1, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Complex of MBD1-MBD and methylated DNA
to be published
|
|
3LVY
 
 | | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Carboxymuconolactone decarboxylase family, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Carboxymuconolactone Decarboxylase Family Protein SMU.961 from Streptococcus mutans
To be Published
|
|
3LS8
 
 | | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol | | Descriptor: | 3-(4-morpholin-4-ylthieno[3,2-d]pyrimidin-2-yl)phenol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Tresaugues, L, Welin, M, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kotenyova, T, Kraulis, P, Moche, M, Nyman, T, Persson, C, Schuler, H, Schutz, P, Siponen, M.I, Thorsell, A.G, Van den Berg, S, Wahlberg, E, Weigelt, J, Wisniewska, M, Nordlund, P, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of human PIK3C3 in complex with 3-[4-(4-Morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol
To be Published
|
|
3LWK
 
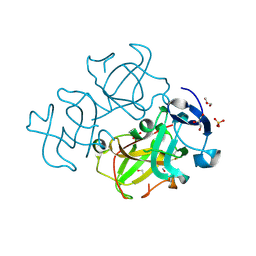 | | Crystal structure of human Beta-crystallin A4 (CRYBA4) | | Descriptor: | Beta-crystallin A4, GLYCEROL, PHOSPHATE ION | | Authors: | Chaikuad, A, Shafqat, N, Krojer, T, Yue, W.W, Cocking, R, Vollmar, M, Muniz, J.R.C, Pike, A.C.W, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Beta-crystallin A4 (CRYBA4)
To be Published
|
|
3MAO
 
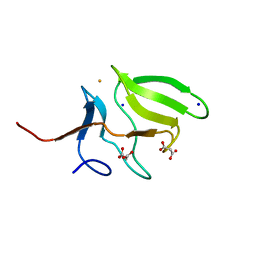 | | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1) | | Descriptor: | FE (III) ION, MALONATE ION, Methionine-R-sulfoxide reductase B1, ... | | Authors: | Chaikuad, A, Shafqat, N, Yue, W.W, Savitsky, P, Krojer, T, Ugochukwu, E, Muniz, J.R.C, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-24 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of Human Methionine-R-Sulfoxide Reductase B1 (MsrB1)
To be Published
|
|
8RU5
 
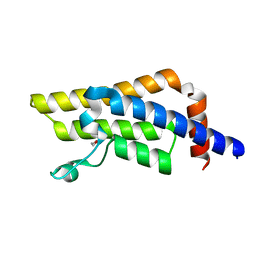 | | ATPase family AAA domain containing 2 with crystallization epitope mutations V1022R:Q1027E | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2 | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-01-30 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
3N73
 
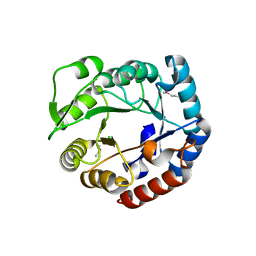 | | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus | | Descriptor: | CHLORIDE ION, Putative 4-hydroxy-2-oxoglutarate aldolase | | Authors: | Cabello, R, Chruszcz, M, Xu, X, Zimmerman, M.D, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-09 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase from Bacillus cereus
To be Published
|
|
3N99
 
 | | Crystal structure of TM1086 | | Descriptor: | CHLORIDE ION, uncharacterized protein TM1086 | | Authors: | Chruszcz, M, Domagalski, M.J, Wang, S, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-28 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Crystal structure of TM1086
To be Published
|
|
2O1Z
 
 | | Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155) | | Descriptor: | FE (III) ION, Ribonucleotide Reductase Subunit R2, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Tempel, W, Qiu, W, Lew, J, Wernimont, A.K, Lin, Y.H, Hassanali, A, Melone, M, Zhao, Y, Nordlund, P, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-11-29 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Plasmodium vivax Ribonucleotide Reductase Subunit R2 (Pv086155)
To be Published
|
|
6GYO
 
 | | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel in complex with cAMP | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOINOSITOL, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Shintre, C.A, Pike, A.C.W, Tessitore, A, Young, M, Bushell, S.R, Strain-Damerell, C, Mukhopadhyay, S, Burgess-Brown, N.A, Huiskonen, J.T, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-30 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of human HCN4 hyperpolarization-activated cyclic nucleotide-gated ion channel
To Be Published
|
|
3NA2
 
 | | Crystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, Uncharacterized protein | | Authors: | Kim, Y, Xu, X, Cui, H, Chin, S, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | Crsystal Structure of Protein of Unknown Function from Mine Drainage Metagenome Leptospirillum rubarum
To be Published
|
|
3NI7
 
 | | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718 | | Descriptor: | Bacterial regulatory proteins, TetR family | | Authors: | Knapik, A, Chruszcz, M, Cymborowski, M, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-06-15 | | Release date: | 2010-08-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal structure of the TetR transcriptional regulator from Nitrosomonas europaea ATCC 19718
To be Published
|
|
3N9X
 
 | | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0 | | Descriptor: | GLYCEROL, Phosphotransferase | | Authors: | Wernimont, A.K, Hutchinson, A, Sullivan, H, MacKenzie, F, Kozieradzki, I, Chau, I, Lew, J, Senisterra, G, Artz, J, Amani, M, Cossar, D, Bochkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Hui, R, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2010-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Map Kinase from plasmodium berghei, PB000659.00.0
To be Published
|
|
2O9X
 
 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | Descriptor: | Reductase, assembly protein | | Authors: | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
6SZM
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2009 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-methyl-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl]piperazine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
2NP5
 
 | | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1. | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, ETHYL DIMETHYL AMMONIO PROPANE SULFONATE, Transcriptional regulator | | Authors: | Chruszcz, M, Evdokimova, E, Kagan, O, Cymborowski, M, Savchenko, A, Edwards, A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-10-26 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a transcriptional regulator (RHA1_ro04179) from Rhodococcus sp. Rha1.
TO BE PUBLISHED
|
|
