1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | Descriptor: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1CM5
 
 | | CRYSTAL STRUCTURE OF C418A,C419A MUTANT OF PFL FROM E.COLI | | Descriptor: | CARBONATE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 1999-12-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
4TLW
 
 | | CARDS TOXIN, FULL-LENGTH | | Descriptor: | ADP-ribosylating toxin CARDS | | Authors: | Becker, A, GALALELDEEN, A, Taylor, A.B, Hart, P.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI IN COMPLEX WITH SUBSTRATE ANALOGUE OXAMATE | | Descriptor: | OXAMIC ACID, PROTEIN (FORMATE ACETYLTRANSFERASE 1) | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-14 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
2VWB
 
 | | Structure of the archaeal Kae1-Bud32 fusion protein MJ1130: a model for the eukaryotic EKC-KEOPS subcomplex involved in transcription and telomere homeostasis. | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PUTATIVE O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Lopreiato, R, Graille, M, Collinet, B, Forterre, P, Domenico, L, van Tilbeurgh, H. | | Deposit date: | 2008-06-20 | | Release date: | 2008-08-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of the Archaeal Kae1/Bud32 Fusion Protein Mj1130: A Model for the Eukaryotic Ekc/Keops Subcomplex
Embo J., 27, 2008
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | Descriptor: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS6
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BSZ
 
 | | PEPTIDE DEFORMYLASE AS FE2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | FE (III) ION, NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1H18
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate | | Descriptor: | FORMATE ACETYLTRANSFERASE 1, L-TREITOL, PYRUVIC ACID, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-04 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1ICJ
 
 | | PDF PROTEIN IS CRYSTALLIZED AS NI2+ CONTAINING FORM, COCRYSTALLIZED WITH INHIBITOR POLYETHYLENE GLYCOL (PEG) | | Descriptor: | NICKEL (II) ION, NONAETHYLENE GLYCOL, PEPTIDE DEFORMYLASE, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-03-12 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
2IVP
 
 | | Structure of UP1 protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE (II) ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2IVN
 
 | | Structure of UP1 protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE, ... | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2IVO
 
 | | Structure of UP1 protein | | Descriptor: | TUNGSTATE(VI)ION, UP1 | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
1H16
 
 | | Pyruvate Formate-Lyase (E.coli) in complex with Pyruvate and CoA | | Descriptor: | COENZYME A, FORMATE ACETYLTRANSFERASE 1, L-TREITOL, ... | | Authors: | Becker, A, Kabsch, W. | | Deposit date: | 2002-07-03 | | Release date: | 2002-11-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structure of Pyruvate Formate-Lyase in Complex with Pyruvate and Coa.How the Enzyme Uses the Cys-418 Thiyl Radical for Pyruvate Cleavage
J.Biol.Chem., 277, 2002
|
|
1H17
 
 | |
2PFL
 
 | | CRYSTAL STRUCTURE OF PFL FROM E.COLI | | Descriptor: | CHLORIDE ION, PROTEIN (PYRUVATE FORMATE-LYASE), SODIUM ION | | Authors: | Becker, A, Fritz-Wolf, K, Kabsch, W, Knappe, J, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1999-05-26 | | Release date: | 1999-12-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and mechanism of the glycyl radical enzyme pyruvate formate-lyase.
Nat.Struct.Biol., 6, 1999
|
|
8CX5
 
 | | Crystal Structure of small molecule alpha,beta-ketoamide 4 covalently bound to K-Ras(G12R) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Morstein, J, Ecker, A, Guiley, K.Z, Shokat, K.M. | | Deposit date: | 2022-05-19 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Chemoselective Covalent Modification of K-Ras(G12R) with a Small Molecule Electrophile.
J.Am.Chem.Soc., 144, 2022
|
|
4GCJ
 
 | | CDK2 in complex with inhibitor RC-3-89 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-amino-5-(2-nitrobenzoyl)-1,3-thiazol-2-yl]amino}benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2012-07-30 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Development of highly potent and selective diaminothiazole inhibitors of cyclin-dependent kinases.
J.Med.Chem., 56, 2013
|
|
3R8L
 
 | | CDK2 in complex with inhibitor L3-4 | | Descriptor: | (5R)-5-tert-butyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QZF
 
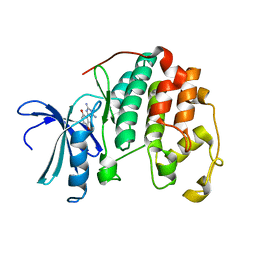 | | CDK2 in complex with inhibitor JWS-6-52 | | Descriptor: | 2-(4,6-diamino-1,3,5-triazin-2-yl)benzene-1,4-diol, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-06 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3R8P
 
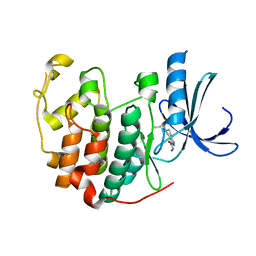 | | CDK2 in complex with inhibitor NSK-MC1-6 | | Descriptor: | (5R)-5-propyl-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-03-24 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3QQL
 
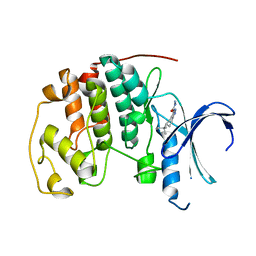 | | CDK2 in complex with inhibitor L3 | | Descriptor: | (5R)-5-(2-methylbutan-2-yl)-4,5,6,7-tetrahydro-1H-indazole-3-carbohydrazide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-02-15 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
3RM6
 
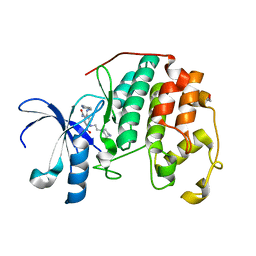 | | CDK2 in complex with inhibitor KVR-2-80 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-aminopyrimidin-5-yl)methyl]amino}-5-nitro-4-{[2-(piperazin-1-yl)ethyl]amino}benzamide, Cyclin-dependent kinase 2 | | Authors: | Betzi, S, Alam, R, Han, H, Becker, A, Schonbrunn, E. | | Deposit date: | 2011-04-20 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-guided optimization of novel CDK2 inhibitors discovered by high-throughput screening
To be Published
|
|
