8QR1
 
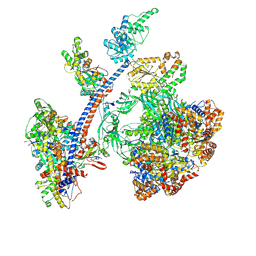 | | Cryo-EM structure of the human Tip60 complex | | Descriptor: | Actin, cytoplasmic 1, N-terminally processed, ... | | Authors: | Li, C, Smirnova, E, Schnitzler, C, Crucifix, C, Concordet, J.P, Brion, A, Poterszman, A, Schultz, P, Papai, G, Ben-Shem, A. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure of human TIP60-C histone exchange and acetyltransferase complex.
Nature, 2024
|
|
4XD4
 
 | | Phosphotriesterase variant E2b | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, Phosphotriesterase variant PTE-R3, ... | | Authors: | Jackson, C.J, Campbell, E, Kaltenbach, M, Tokuriki, N. | | Deposit date: | 2014-12-19 | | Release date: | 2015-12-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The role of protein dynamics in the evolution of new enzyme function.
Nat.Chem.Biol., 12, 2016
|
|
7QYF
 
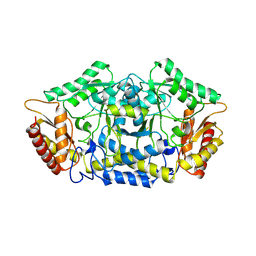 | | Structure of the transaminase PluriZyme variant (TR2E2) | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-07-26 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
7QYG
 
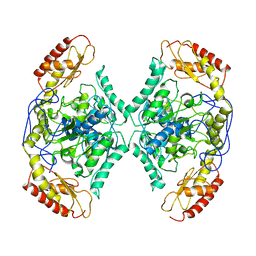 | | Structure of the transaminase TR2 | | Descriptor: | Aminotransferase TR2 | | Authors: | Roda, S, Fernandez-Lopez, L, Benedens, M, Bollinger, A, Thies, S, Schumacher, J, Coscolin, C, Kazemi, M, Santiago, G, Gertzen, C.G, Gonzalez-Alfonso, J, Plou, F.J, Jaeger, K.E, Smits, S.H, Ferrer, M, Guallar, V. | | Deposit date: | 2022-01-28 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A Plurizyme with Transaminase and Hydrolase Activity Catalyzes Cascade Reactions.
Angew Chem Int Ed Engl, 61, 2022
|
|
4XQD
 
 | | X-ray structure analysis of xylanase-WT at pH4.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Endo-1,4-beta-xylanase 2, IODIDE ION | | Authors: | Wan, Q, Park, J.M, Riccardi, D.M, Hanson, L.B, Fisher, Z, Smith, J.C, Ostermann, A, Schrader, T, Graham, D.E, Coates, L, Langan, P, Kovalevsky, A.Y. | | Deposit date: | 2015-01-19 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Direct determination of protonation states and visualization of hydrogen bonding in a glycoside hydrolase with neutron crystallography.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3NQU
 
 | |
5OCD
 
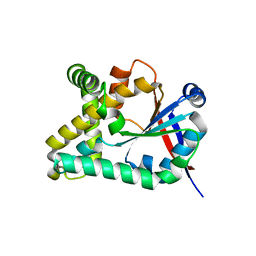 | |
8ANW
 
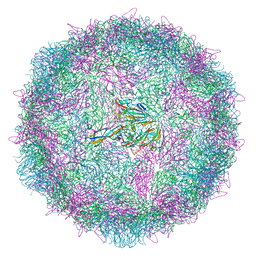 | | Poliovirus type 3 (strain Saukett) stabilised virus-like particle (PV3 SC8). | | Descriptor: | Capsid protein, VP0, VP1, ... | | Authors: | Bahar, M.W, Fry, E.E, Stuart, D.I. | | Deposit date: | 2022-08-06 | | Release date: | 2022-10-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Production and Characterisation of Stabilised PV-3 Virus-like Particles Using Pichia pastoris .
Viruses, 14, 2022
|
|
1B97
 
 | |
6R8P
 
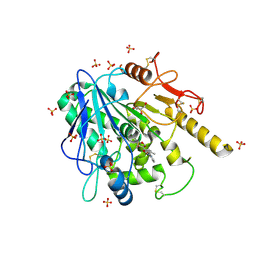 | | Notum fragment 723 | | Descriptor: | 2-(2-methylphenoxy)-~{N}-pyridin-3-yl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
1B79
 
 | |
6CLV
 
 | | Staphylococcus aureus Dihydropteroate Synthase (saDHPS) F17L E208K double mutant structure | | Descriptor: | 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-N-(3,4-dimethyl-1,2-oxazol-5-yl)benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Gajewski, S, Griffith, E.C, Wu, Y, White, S.W. | | Deposit date: | 2018-03-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural and Functional Basis for Recurring Sulfa Drug Resistance Mutations inStaphylococcus aureusDihydropteroate Synthase.
Front Microbiol, 9, 2018
|
|
8CTI
 
 | | Cryo-EM structure of human METTL1-WDR4-tRNA(Val) complex | | Descriptor: | tRNA (guanine-N(7)-)-methyltransferase, tRNA (guanine-N(7)-)-methyltransferase non-catalytic subunit WDR4, tRNA-Val-TAC-2-1 | | Authors: | Li, J, Wang, L, Fontana, P, Hunkeler, M, Roy-Burman, S.S, Wu, H, Fischer, E.S, Gregory, R.I. | | Deposit date: | 2022-05-14 | | Release date: | 2022-12-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of regulated m 7 G tRNA modification by METTL1-WDR4.
Nature, 613, 2023
|
|
5W3F
 
 | | Yeast tubulin polymerized with GTP in vitro | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Howes, S.C, Geyer, E.A, LaFrance, B, Zhang, R, Kellogg, E.H, Westermann, S, Rice, L.M, Nogales, E. | | Deposit date: | 2017-06-07 | | Release date: | 2017-07-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural differences between yeast and mammalian microtubules revealed by cryo-EM.
J. Cell Biol., 216, 2017
|
|
1AVC
 
 | |
5OAB
 
 | | A novel crystal form of human RNase6 at atomic resolution | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, POTASSIUM ION, ... | | Authors: | Prats-Ejarque, G, Moussaoui, M, Boix, E. | | Deposit date: | 2017-06-21 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.111 Å) | | Cite: | Characterization of an RNase with two catalytic centers. Human RNase6 catalytic and phosphate-binding site arrangement favors the endonuclease cleavage of polymeric substrates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
2AHU
 
 | | Crystal structure of Acyl-CoA transferase (YdiF) apoenzyme from Escherichia coli O157:H7. | | Descriptor: | putative enzyme ydiF | | Authors: | Rangarajan, E.S, Li, Y, Ajamian, E, Iannuzzi, P, Kernaghan, S.D, Fraser, M.E, Cygler, M, Matte, A, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2005-07-28 | | Release date: | 2005-11-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic trapping of the glutamyl-CoA thioester intermediate of family I CoA transferases.
J.Biol.Chem., 280, 2005
|
|
5DWG
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Triaryl-substituted Imine Analog, 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol | | Descriptor: | 4-{(E)-(4-hydroxyphenyl)[(2-methylphenyl)imino]methyl}benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6TQJ
 
 | | Crystal structure of the c14 ring of the F1FO ATP synthase from spinach chloroplast | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, ATP synthase subunit c, chloroplastic, ... | | Authors: | Kovalev, K, Gushchin, I, Vlasov, A, Round, E, Polovinkin, V, Gordeliy, V. | | Deposit date: | 2019-12-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unusual features of the c-ring of F1FOATP synthases.
Sci Rep, 9, 2019
|
|
3HV0
 
 | |
6HKW
 
 | | Crystal structure of human SDS22 | | Descriptor: | Protein phosphatase 1 regulatory subunit 7, SULFATE ION | | Authors: | Heroes, E, Choy, M.S, Page, R, Peti, W, Ulens, C, Van Meervelt, L, Nys, M, Bollen, M. | | Deposit date: | 2018-09-09 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structure-Guided Exploration of SDS22 Interactions with Protein Phosphatase PP1 and the Splicing Factor BCLAF1.
Structure, 27, 2019
|
|
5OJM
 
 | | Structure of a chimaeric beta3-alpha5 GABAA receptor in complex with nanobody Nb25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Human GABAA receptor chimera beta3-alpha5,Gamma-aminobutyric acid receptor subunit beta-3,Gamma-aminobutyric acid receptor subunit alpha-5, Nanobody Nb25, ... | | Authors: | Miller, P.S, Scott, S, Masiulis, S, De Colibus, L, Pardon, E, Steyaert, J, Aricescu, A.R. | | Deposit date: | 2017-07-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for GABAA receptor potentiation by neurosteroids.
Nat. Struct. Mol. Biol., 24, 2017
|
|
1SE7
 
 | | Solution structure of the E. coli bacteriophage P1 encoded HOT protein: a homologue of the theta subunit of E. coli DNA polymerase III | | Descriptor: | HOMOLOGUE OF THE THETA SUBUNIT OF DNA POLYMERASE III | | Authors: | DeRose, E.F, Kirby, T.W, Mueller, G.A, Chikova, A.K, Schaaper, R.M, London, R.E. | | Deposit date: | 2004-02-16 | | Release date: | 2004-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Phage Like It HOT: Solution Structure of the Bacteriophage P1-Encoded HOT Protein, a Homolog of the theta Subunit of E. coli DNA Polymerase III
Structure, 12, 2004
|
|
1BI9
 
 | | RETINAL DEHYDROGENASE TYPE TWO WITH NAD BOUND | | Descriptor: | CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, RETINAL DEHYDROGENASE TYPE II | | Authors: | Newcomer, M.E, Lamb, A.L. | | Deposit date: | 1998-06-23 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of retinal dehydrogenase type II at 2.7 A resolution: implications for retinal specificity.
Biochemistry, 38, 1999
|
|
6ZRF
 
 | |
