6D7G
 
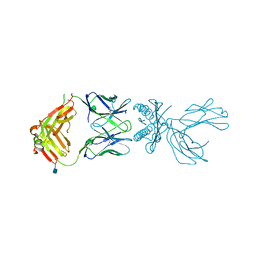 | | Structure of 5F3 TCR in complex with HLA-A2/MART-1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MART1 PEPTIDE-BETA-2-MICROGLOBULIN-HLA-A*02 CHIMERA, T-CELL RECEPTOR GAMMA VARIABLE 8,T-CELL RECEPTOR GAMMA-2 CHAIN C REGION, ... | | Authors: | Roy, S, Adams, E.J. | | Deposit date: | 2018-04-24 | | Release date: | 2019-01-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Generation and molecular recognition of melanoma-associated antigen-specific human gamma delta T cells.
Sci Immunol, 3, 2018
|
|
7KRN
 
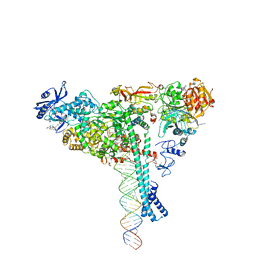 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KKW
 
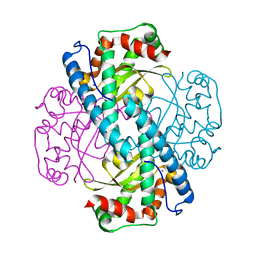 | | Neutron structure of Reduced Human MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial, ... | | Authors: | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | Deposit date: | 2020-10-28 | | Release date: | 2021-04-21 | | Last modified: | 2024-04-10 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
7KRP
 
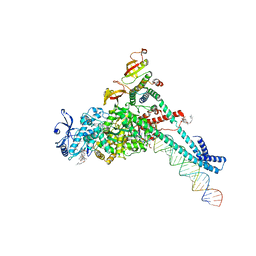 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPSO, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRO
 
 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
2WS3
 
 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhD Tyr83Phe mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-09-03 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Succinate Dehydrogenase Activity
To be Published
|
|
1URA
 
 | | ALKALINE PHOSPHATASE (D51ZN) | | Descriptor: | ALKALINE PHOSPHATASE, PHOSPHATE ION, ZINC ION | | Authors: | Tibbitts, T.T, Murphy, J.E, Kantrowitz, E.R. | | Deposit date: | 1996-02-03 | | Release date: | 1996-07-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Kinetic and structural consequences of replacing the aspartate bridge by asparagine in the catalytic metal triad of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 257, 1996
|
|
1H0M
 
 | | Three-dimensional structure of the quorum sensing protein TraR bound to its autoinducer and to its target DNA | | Descriptor: | 3-OXO-OCTANOIC ACID (2-OXO-TETRAHYDRO-FURAN-3-YL)-AMIDE, 5'-D(*AP*TP*GP*TP*GP*CP*AP*GP*AP*TP *CP*TP*GP*CP*AP*CP*AP*T)-3', Transcriptional activator protein TraR | | Authors: | Vannini, A, Volpari, C, Gargioli, C, Muraglia, E, Cortese, R, De Francesco, R, Neddermann, P, Di Marco, S. | | Deposit date: | 2002-06-25 | | Release date: | 2002-08-29 | | Last modified: | 2020-12-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Crystal Structure of the Quorum Sensing Protein Trar Bound to its Autoinducer and Target DNA
Embo J., 21, 2002
|
|
1D0A
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 (TRAF2) IN COMPLEX WITH A HUMAN OX40 PEPTIDE | | Descriptor: | OX40L RECEPTOR PEPTIDE, TUMOR NECROSIS FACTOR RECEPTOR ASSOCIATED PROTEIN 2 | | Authors: | Ye, H, Park, Y.C, Kreishman, M, Kieff, E, Wu, H. | | Deposit date: | 1999-09-09 | | Release date: | 2000-03-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the recognition of diverse receptor sequences by TRAF2.
Mol.Cell, 4, 1999
|
|
2WU2
 
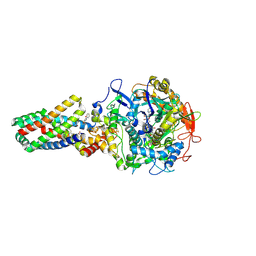 | | Crystal structure of the E. coli succinate:quinone oxidoreductase (SQR) SdhC His84Met mutant | | Descriptor: | 2-METHYL-N-PHENYL-5,6-DIHYDRO-1,4-OXATHIINE-3-CARBOXAMIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Ruprecht, J, Yankovskaya, V, Maklashina, E, Iwata, S, Cecchini, G. | | Deposit date: | 2009-09-28 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the E. Coli Succinate:Quinone Oxidoreductase (Sqr) Sdhc His84met Mutant
To be Published
|
|
1CLI
 
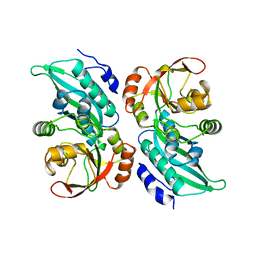 | | X-RAY CRYSTAL STRUCTURE OF AMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE (PURM), FROM THE E. COLI PURINE BIOSYNTHETIC PATHWAY, AT 2.5 A RESOLUTION | | Descriptor: | PROTEIN (PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE), SULFATE ION | | Authors: | Li, C, Kappock, T.J, Stubbe, J, Weaver, T.M, Ealick, S.E. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
Structure Fold.Des., 7, 1999
|
|
1MCZ
 
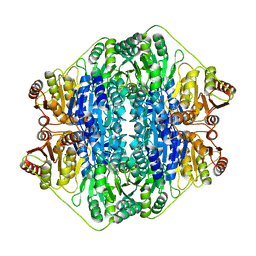 | | BENZOYLFORMATE DECARBOXYLASE FROM PSEUDOMONAS PUTIDA COMPLEXED WITH AN INHIBITOR, R-MANDELATE | | Descriptor: | (R)-MANDELIC ACID, BENZOYLFORMATE DECARBOXYLASE, MAGNESIUM ION, ... | | Authors: | Polovnikova, E.S, Bera, A.K, Hasson, M.S. | | Deposit date: | 2002-08-06 | | Release date: | 2003-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and Kinetic Analysis of Catalysis by a Thiamin
Diphosphate-Dependent Enzyme, Benzoylformate Decarboxylase
Biochemistry, 42, 2003
|
|
1D0J
 
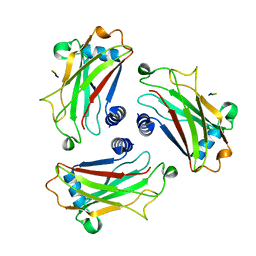 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 IN COMPLEX WITH A M4-1BB PEPTIDE | | Descriptor: | 4-1BB LIGAND RECEPTOR, TUMOR NECROSIS FACTOR RECEPTOR ASSOCIATED PROTEIN 2 | | Authors: | Ye, H, Park, Y.C, Kreishman, M, Kieff, E, Wu, H. | | Deposit date: | 1999-09-10 | | Release date: | 2000-03-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis for the recognition of diverse receptor sequences by TRAF2.
Mol.Cell, 4, 1999
|
|
1CF4
 
 | | CDC42/ACK GTPASE-BINDING DOMAIN COMPLEX | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (ACTIVATED P21CDC42HS KINASE), ... | | Authors: | Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Lim, L, Laue, E.D. | | Deposit date: | 1999-03-23 | | Release date: | 1999-06-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the small G protein Cdc42 bound to the GTPase-binding domain of ACK.
Nature, 399, 1999
|
|
3KDD
 
 | | Crystal Structure of HIV-1 Protease (Q7K, L33I, L63I) in Complex with KNI-10265 | | Descriptor: | (4R)-3-[(2S,3S)-3-{[(2,6-difluorophenoxy)acetyl]amino}-2-hydroxy-4-phenylbutanoyl]-N-[(1S,2R)-2-hydroxy-2,3-dihydro-1H- inden-1-yl]-5,5-dimethyl-1,3-thiazolidine-4-carboxamide, GLYCEROL, Protease | | Authors: | Chufan, E.E, Kawasaki, Y, Freire, E, Amzel, L.M. | | Deposit date: | 2009-10-22 | | Release date: | 2010-03-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | How much binding affinity can be gained by filling a cavity?
Chem.Biol.Drug Des., 75, 2010
|
|
2OVQ
 
 | | Structure of the Skp1-Fbw7-CyclinEdegC complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A, SULFATE ION, ... | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
5C83
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 21 | | Descriptor: | (2R,5R)-4-[2-(6-benzyl-3,3-dimethyl-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-5-(methoxymethyl)-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-25 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
1CZZ
 
 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 IN COMPLEX WITH A 17-RESIDUE CD40 PEPTIDE | | Descriptor: | CD 40 PEPTIDE, TUMOR NECROSIS FACTOR RECEPTOR ASSOCIATED PROTEIN 2 | | Authors: | Ye, H, Park, Y.C, Kreishman, M, Kieff, E, Wu, H. | | Deposit date: | 1999-09-07 | | Release date: | 2000-03-08 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for the recognition of diverse receptor sequences by TRAF2.
Mol.Cell, 4, 1999
|
|
1CMC
 
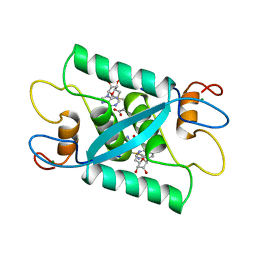 | |
1Z0E
 
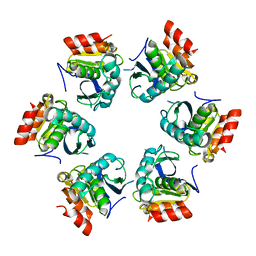 | | Crystal Structure of A. fulgidus Lon proteolytic domain | | Descriptor: | Putative protease La homolog type | | Authors: | Botos, I, Melnikov, E.E, Cherry, S, Kozlov, S, Makhovskaya, O.V, Tropea, J.E, Gustchina, A, Rotanova, T.V, Wlodawer, A. | | Deposit date: | 2005-03-01 | | Release date: | 2005-08-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Atomic-resolution Crystal Structure of the Proteolytic Domain of Archaeoglobus fulgidus Lon Reveals the Conformational Variability in the Active Sites of Lon Proteases
J.Mol.Biol., 351, 2005
|
|
1J2O
 
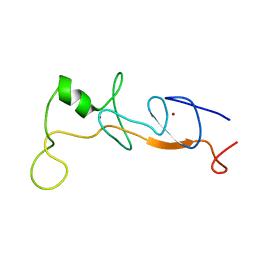 | | Structure of FLIN2, a complex containing the N-terminal LIM domain of LMO2 and ldb1-LID | | Descriptor: | Fusion of Rhombotin-2 and LIM domain-binding protein 1, ZINC ION | | Authors: | Deane, J.E, Mackay, J.P, Kwan, A.H, Sum, E.Y, Visvader, J.E, Matthews, J.M. | | Deposit date: | 2003-01-08 | | Release date: | 2003-05-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recognition of ldb1 by the N-terminal LIM domains of LMO2 and LMO4
EMBO J., 22, 2003
|
|
1D01
 
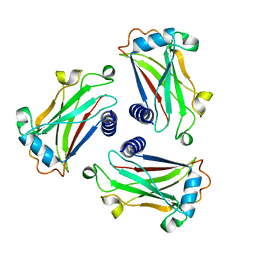 | | STRUCTURE OF TNF RECEPTOR ASSOCIATED FACTOR 2 IN COMPLEX WITH A HUMAN CD30 PEPTIDE | | Descriptor: | CD30 PEPTIDE, TUMOR NECROSIS FACTOR RECEPTOR ASSOCIATED FACTOR 2 | | Authors: | Ye, H, Park, Y.C, Kreishman, M, Kieff, E, Wu, H. | | Deposit date: | 1999-09-07 | | Release date: | 2003-12-02 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for the recognition of diverse receptor sequences by TRAF2.
Mol.Cell, 4, 1999
|
|
3KU4
 
 | | Trapping of an oxocarbenium ion intermediate in UP crystals | | Descriptor: | SULFATE ION, Uridine phosphorylase | | Authors: | Paul, D, O'Leary, S, Rajashankar, K, Bu, W, Toms, A, Settembre, E, Sanders, J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2009-11-26 | | Release date: | 2010-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Glycal formation in crystals of uridine phosphorylase.
Biochemistry, 49, 2010
|
|
1YZ9
 
 | | Crystal structure of RNase III mutant E110Q from Aquifex aeolicus complexed with double stranded RNA at 2.1-Angstrom Resolution | | Descriptor: | 5'-R(*CP*GP*AP*AP*CP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III, SULFATE ION | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-28 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
1HBS
 
 | |
