7OEB
 
 | | Lassa virus L protein bound to 3' promoter RNA (well-resolved endonuclease) [3END-ENDO] | | Descriptor: | 3' vRNA, MAGNESIUM ION, RNA-directed RNA polymerase L, ... | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-02 | | Release date: | 2021-12-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
7AIF
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND L-GLUTAMATE TENOFOVIR WITH BOUND MANGANESE | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), Gag-Pol polyprotein, ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-27 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7AID
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEX WITH DNA AND D-ASPARTATE TENOFOVIR | | Descriptor: | D-Aspartate Tenofovir, DNA (5'-D(*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(P*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Gu, W, Martinez, S.E, Nguyen, H, Xu, H, Herdewijn, P, de Jonghe, S, Das, K. | | Deposit date: | 2020-09-26 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tenofovir-Amino Acid Conjugates Act as Polymerase Substrates-Implications for Avoiding Cellular Phosphorylation in the Discovery of Nucleotide Analogues.
J.Med.Chem., 64, 2021
|
|
7QI3
 
 | | Structure of Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase | | Descriptor: | 1,2-ETHANEDIOL, Arylamine N-acetyltransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lowe, E.D, Kotomina, E, Karagianni, E, Boukouvala, S. | | Deposit date: | 2021-12-14 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fusarium verticillioides NAT1 (FDB2) N-malonyltransferase is structurally, functionally and phylogenetically distinct from its N-acetyltransferase (NAT) homologues.
Febs J., 290, 2023
|
|
7ON9
 
 | | Crystal structure of para-hydroxybenzoate-3-hydroxylase PraI | | Descriptor: | 4-hydroxybenzoate 3-monooxygenase (NAD(P)H), FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID | | Authors: | Zahn, M, McGeehan, J.E. | | Deposit date: | 2021-05-25 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Debottlenecking 4-hydroxybenzoate hydroxylation in Pseudomonas putida KT2440 improves muconate productivity from p-coumarate.
Metab Eng, 70, 2022
|
|
8OET
 
 | | SFX structure of the class II photolyase complexed with a thymine dimer | | Descriptor: | DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, DNA (14-mer), Deoxyribodipyrimidine photo-lyase, ... | | Authors: | Lane, T.J, Christou, N.-E, Melo, D.V.M, Apostolopoulou, V, Pateras, A, Mashhour, A.R, Galchenkova, M, Gunther, S, Reinke, P, Kremling, V, Oberthuer, D, Henkel, A, Sprenger, J, Scheer, T.E.S, Lange, E, Yefanov, O.N, Middendorf, P, Sellberg, J.A, Schubert, R, Fadini, A, Cirelli, C, Beale, E.V, Johnson, P, Dworkowski, F, Ozerov, D, Bertrand, Q, Wranik, M, Zitter, E.D, Turk, D, Bajt, S, Chapman, H, Bacellar, C. | | Deposit date: | 2023-03-12 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Time-resolved crystallography captures light-driven DNA repair.
Science, 382, 2023
|
|
8CII
 
 | | Delta-RBD complex with BA.2-07 fab, SARS1-34 fab and C1 nanobody | | Descriptor: | BA.2-07 fab Heavy Chain, BA.2-07 fab Light Chain, C1 nanobody, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-09 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Potent cross-reactive mAbs from BA.4/5 breakthrough infection
To Be Published
|
|
7B62
 
 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
7OPT
 
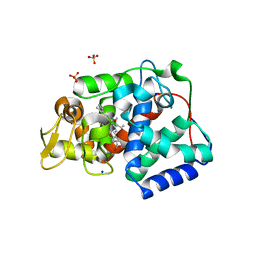 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | Ascorbate peroxidase, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-01 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
8CXM
 
 | | Cryo-EM structure of the supercoiled E. coli K12 flagellar filament core, Normal waveform | | Descriptor: | Flagellin | | Authors: | Sonani, R.R, Kreutzberger, M.A.B, Sebastian, A.L, Scharf, B, Egelman, E.H. | | Deposit date: | 2022-05-21 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Convergent evolution in the supercoiling of prokaryotic flagellar filaments.
Cell, 185, 2022
|
|
7OQR
 
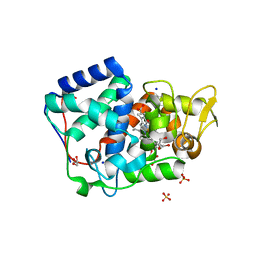 | | Crystal structure of Trypanosoma cruzi peroxidase | | Descriptor: | ACETATE ION, Ascorbate peroxidase, GLYCEROL, ... | | Authors: | Freeman, S.L, Kwon, H, Skafar, V, Fielding, A.J, Martinez, A, Piacenza, L, Radi, R, Raven, E.L. | | Deposit date: | 2021-06-04 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of Trypanosoma cruzi heme peroxidase and characterization of its substrate specificity and compound I intermediate.
J.Biol.Chem., 298, 2022
|
|
6GGM
 
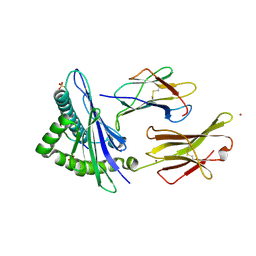 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P2-Phe. | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Mtb44*P2-Phe peptide variant (ARG-PHE-PRO-ALA-LYS-ALA-PRO-LEU-LEU), ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
6GH1
 
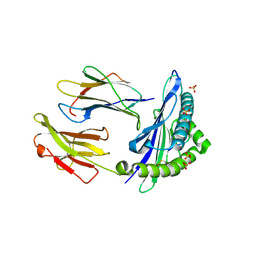 | | HLA-E*01:03 in complex with Mtb44 | | Descriptor: | Beta-2-microglobulin, Enoyl-[acyl-carrier-protein] reductase [NADH], MHC class I antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-04 | | Release date: | 2018-08-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
8D7U
 
 | |
8D81
 
 | | Cereblon~DDB1 bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8CVJ
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-NH-tRNAphe, peptidyl P-site fMSEAC-NH-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-05-18 | | Release date: | 2022-10-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.
Nat.Chem., 15, 2023
|
|
8CVK
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-NH-tRNAphe, peptidyl P-site fMRC-NH-tRNAmet, and deacylated E-site tRNAphe at 2.50A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-05-18 | | Release date: | 2022-10-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.
Nat.Chem., 15, 2023
|
|
8CVP
 
 | | Cereblon-DDB1 in the Apo form | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, ZINC ION | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8D7V
 
 | |
8D7W
 
 | |
8D7Z
 
 | | Cereblon-DDB1 bound to CC-92480 and Ikaros ZF1-2-3 | | Descriptor: | DNA damage-binding protein 1, DNA-binding protein Ikaros, Mezigdomide, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
8CVL
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Phe-NH-tRNAphe, peptidyl P-site fMTHSMRC-NH-tRNAmet, and deacylated E-site tRNAphe at 2.30A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-05-18 | | Release date: | 2022-10-19 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the ribosome function from the structures of non-arrested ribosome-nascent chain complexes.
Nat.Chem., 15, 2023
|
|
8D80
 
 | | Cereblon~DDB1 bound to Iberdomide and Ikaros ZF1-2-3 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, DNA-binding protein Ikaros, ... | | Authors: | Watson, E.R, Lander, G.C. | | Deposit date: | 2022-06-07 | | Release date: | 2022-07-20 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular glue CELMoD compounds are regulators of cereblon conformation.
Science, 378, 2022
|
|
6GHN
 
 | | HLA-E*01:03 in complex with the Mtb44 peptide variant: Mtb44*P9-Phe. | | Descriptor: | ARG-LEU-PRO-ALA-LYS-ALA-PRO-LEU-PHE, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Walters, L.C, Gillespie, G.M, McMichael, A.J, Rozbesky, D, Jones, E.Y, Harlos, K. | | Deposit date: | 2018-05-08 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.542 Å) | | Cite: | Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding.
Nat Commun, 9, 2018
|
|
8CIN
 
 | | BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-5 fab HEAVY CHAIN, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
