8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
5I1N
 
 | | Villin headpiece subdomain with a Gln26 to beta-3-homoglutamine substitution | | Descriptor: | D-Villin headpiece subdomain, Villin-1 | | Authors: | Kreitler, D.F, Mortenson, D.E, Gellman, S.H, Forest, K.T. | | Deposit date: | 2016-02-05 | | Release date: | 2016-05-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effects of Single alpha-to-beta Residue Replacements on Structure and Stability in a Small Protein: Insights from Quasiracemic Crystallization.
J.Am.Chem.Soc., 138, 2016
|
|
7YXA
 
 | | XFEL crystal structure of the human sphingosine 1 phosphate receptor 5 in complex with ONO-5430608 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[6-(2-naphthalen-1-ylethoxy)-2,3,4,5-tetrahydro-1H-3-benzazepin-3-ium-3-yl]butanoic acid, ... | | Authors: | Lyapina, E, Marin, E, Gusach, A, Orekhov, P, Gerasimov, A, Luginina, A, Vakhrameev, D, Ergasheva, M, Kovaleva, M, Khusainov, G, Khorn, P, Shevtsov, M, Kovalev, K, Okhrimenko, I, Bukhdruker, S, Popov, P, Hu, H, Weierstall, U, Liu, W, Cho, Y, Gushchin, I, Rogachev, A, Bourenkov, G, Park, S, Park, G, Huyn, H.J, Park, J, Gordeliy, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2022-02-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for receptor selectivity and inverse agonism in S1P 5 receptors.
Nat Commun, 13, 2022
|
|
4X8J
 
 | | Crystal Structure of murine 12F4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | 12F4 FAB Heavy chain, 12F4 FAB Light chain, NONAETHYLENE GLYCOL, ... | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-04-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
5EEY
 
 | | RADIATION DAMAGE TO THE TRAP-RNA COMPLEX: DOSE (DWD) 11.6 MGy | | Descriptor: | (GAGUU)10GAG 53-NUCLEOTIDE RNA, TRYPTOPHAN, Transcription attenuation protein MtrB | | Authors: | Bury, C.S, McGeehan, J.E, Garman, E.F, Shevtsov, M.B. | | Deposit date: | 2015-10-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | RNA protects a nucleoprotein complex against radiation damage.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8AXS
 
 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
8AXI
 
 | | Sialidases and Fucosidases of Akkermansia muciniphila are key for rapid growth on colonic mucin and nutrient sharing amongst mucin-associated human gut microbiota | | Descriptor: | 1,2-ETHANEDIOL, 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, CALCIUM ION, ... | | Authors: | Sakanaka, H, Nielsen, T.S, Pichler, M.J, Nordberg Karlsson, E, Abou Hachem, M, Morth, J.P. | | Deposit date: | 2022-08-31 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Sialidases and fucosidases of Akkermansia muciniphila are crucial for growth on mucin and nutrient sharing with mucus-associated gut bacteria.
Nat Commun, 14, 2023
|
|
7Z20
 
 | | 70S E. coli ribosome with an extended uL23 loop from Candidatus marinimicrobia and a stalled filamin domain 5 nascent chain | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Mitropoulou, A, Plessa, E, Wlodarski, T, Ahn, M, Sidhu, H, Becker, T.A, Beckmann, R, Cabrita, L.D, Christodoulou, J. | | Deposit date: | 2022-02-25 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Modulating co-translational protein folding by rational design and ribosome engineering.
Nat Commun, 13, 2022
|
|
6BVE
 
 | |
9B9V
 
 | |
4X2C
 
 | | Clostridium difficile Fic protein_0569 mutant S31A, E35A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fic family protein putative filamentation induced by cAMP protein, GLYCEROL, ... | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Clostridium difficile Fic_0569 S31A, E35A mutant at 1.8 Angstroms resolution
To Be Published
|
|
8AWI
 
 | | Crystal structure of Human Transthyretin at 1.15 Angstrom resolution | | Descriptor: | SODIUM ION, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2022-08-29 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
6N74
 
 | | Crystal Structure of ATPase delta1-79 Spa47 R271E | | Descriptor: | ATP synthase SpaL/MxiB, SULFATE ION | | Authors: | Morales, Y, Johnson, S.J, Demler, H.J, Dickenson, N.E. | | Deposit date: | 2018-11-27 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Interfacial amino acids support Spa47 oligomerization and shigella type three secretion system activation.
Proteins, 87, 2019
|
|
4WR7
 
 | | Crystal structure of human carbonic anhydrase isozyme I with 2,3,5,6-Tetrafluoro-4-(propylthio)benzenesulfonamide. | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-(propylsulfanyl)benzenesulfonamide, ACETATE ION, ... | | Authors: | Manakova, E, Smirnov, A, Grazulis, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Intrinsic thermodynamics of 4-substituted-2,3,5,6-tetrafluorobenzenesulfonamide binding to carbonic anhydrases by isothermal titration calorimetry.
Biophys.Chem., 205, 2015
|
|
6PKF
 
 | | Myocilin OLF mutant N428E/D478K | | Descriptor: | GLYCEROL, Myocilin | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.484 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6BWD
 
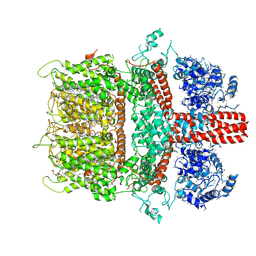 | | 3.7 angstrom cryoEM structure of truncated mouse TRPM7 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, MAGNESIUM ION, Transient receptor potential cation channel subfamily M member 7 | | Authors: | Zhang, J, Li, Z, Duan, J, Li, J, Hulse, R.E, Santa-Cruz, A, Abiria, S.A, Krapivinsky, G, Clapham, D.E. | | Deposit date: | 2017-12-14 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure of the mammalian TRPM7, a magnesium channel required during embryonic development.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C1H
 
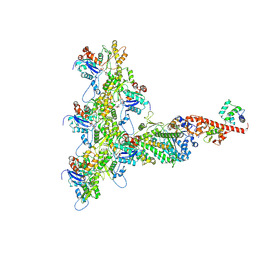 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8HUE
 
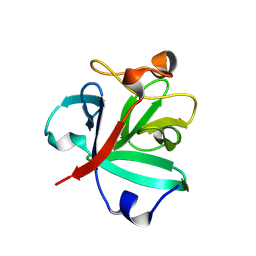 | | Crystal structure of FGF2-M2 mutant - D28E/C78I/C96I/S137P | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Fibroblast growth factor 2 | | Authors: | Jung, Y.E, Cha, S.S, An, Y.J. | | Deposit date: | 2022-12-23 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural and biochemical investigation into stable FGF2 mutants with novel mutation sites and hydrophobic replacements for surface-exposed cysteines.
Plos One, 19, 2024
|
|
6QVL
 
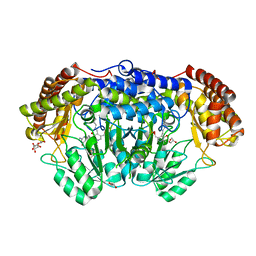 | | Human SHMT2 in complex with pemetrexed | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Scaletti, E, Jemth, A.S, Helleday, T, Stenmark, P. | | Deposit date: | 2019-03-03 | | Release date: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural basis of inhibition of the human serine hydroxymethyltransferase SHMT2 by antifolate drugs.
Febs Lett., 593, 2019
|
|
7Z5U
 
 | | Crystal structure of the peptidase domain of collagenase G from Clostridium histolyticum in complex with a hydroxamate-based inhibitor | | Descriptor: | (2~{R})-~{N}-[2-[4-[(2-acetamidophenoxy)methyl]-1,2,3-triazol-1-yl]ethyl]-2-(2-methylpropyl)-~{N}'-oxidanyl-propanediamide, CALCIUM ION, Collagenase ColG, ... | | Authors: | Schoenauer, E, Brandstetter, H. | | Deposit date: | 2022-03-10 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery and Characterization of Synthesized and FDA-Approved Inhibitors of Clostridial and Bacillary Collagenases.
J.Med.Chem., 65, 2022
|
|
5WJY
 
 | |
6MLQ
 
 | | Cryo-EM structure of microtubule-bound Kif7 in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
6QZL
 
 | | Structure of the H1 domain of human KCTD12 | | Descriptor: | BTB/POZ domain-containing protein KCTD12 | | Authors: | Pinkas, D.M, Bufton, J.C, Fox, A.E, Newman, J.A, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-11 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the H1 domain of human KCTD12
To be published
|
|
6MM9
 
 | | Diheteromeric NMDA receptor GluN1/GluN2A in the '1-Knuckle' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 6.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-29 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6C29
 
 | | Crystal structure of the N-terminal periplasmic domain of ScsB from Proteus mirabilis | | Descriptor: | Putative metal resistance protein | | Authors: | Furlong, E.J, Choudhury, H.G, Kurth, F, Martin, J.L. | | Deposit date: | 2018-01-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.538 Å) | | Cite: | Disulfide isomerase activity of the dynamic, trimericProteus mirabilisScsC protein is primed by the tandem immunoglobulin-fold domain of ScsB.
J. Biol. Chem., 293, 2018
|
|
