1KG5
 
 | | Crystal structure of the K142Q mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
4FL4
 
 | | Scaffoldin conformation and dynamics revealed by a ternary complex from the Clostridium thermocellum cellulosome | | Descriptor: | CALCIUM ION, Cellulosome anchoring protein cohesin region, Glycoside hydrolase family 9, ... | | Authors: | Currie, M.A, Adams, J.J, Faucher, F, Bayer, E.A, Jia, Z, Smith, S.P, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Scaffoldin Conformation and Dynamics Revealed by a Ternary Complex from the Clostridium thermocellum Cellulosome.
J.Biol.Chem., 287, 2012
|
|
6CWD
 
 | |
5KR0
 
 | | Protease E35D-APV | | Descriptor: | GLYCEROL, Protease E35D-APV, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Liu, Z, Poole, K.M, Mahon, B.P, McKenna, R, Fanucci, G.E. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-21 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Effects of Hinge-region Natural Polymorphisms on Human Immunodeficiency Virus-Type 1 Protease Structure, Dynamics, and Drug Pressure Evolution.
J.Biol.Chem., 291, 2016
|
|
6GMP
 
 | | CRYSTAL STRUCTURE OF THE PPIASE DOMAIN OF TBPAR42 | | Descriptor: | PARVULIN 42 | | Authors: | Hoenig, D, Rute, A, Hofmann, E, Bayer, P, Gasper, R. | | Deposit date: | 2018-05-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Analysis of the 42 kDa Parvulin ofTrypanosoma brucei.
Biomolecules, 9, 2019
|
|
5A23
 
 | | SdsA sulfatase triclinic form | | Descriptor: | SDS HYDROLASE SDSA1, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncik, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-05-11 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
2OVP
 
 | | Structure of the Skp1-Fbw7 complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
5ACJ
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
6GXS
 
 | | Crystal structure of CV39L lectin from Chromobacterium violaceum at 1.8 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CV39L lectin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sykorova, P, Novotna, J, Demo, G, Pompidor, G, Dubska, E, Komarek, J, Fujdiarova, E, Haronikova, L, Varrot, A, Imberty, A, Shilova, N, Bovin, N, Pokorna, M, Wimmerova, M. | | Deposit date: | 2018-06-27 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of novel lectins from Burkholderia pseudomallei and Chromobacterium violaceum with seven-bladed beta-propeller fold.
Int.J.Biol.Macromol., 152, 2020
|
|
5DKR
 
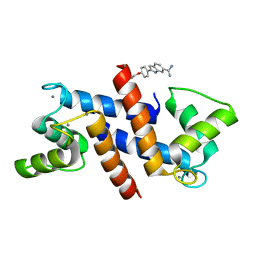 | | Crystal Structure of Calcium-loaded S100B bound to SBi29 | | Descriptor: | 2-[4-(4-carbamimidoylphenoxy)phenyl]-1H-indole-6-carboximidamide, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Ansari, M.I, Pierce, A.D, Wilder, P.T, McKnight, L.E, Raman, E.P, Neau, D.B, Bezawada, P, Alasady, M.J, Varney, K.M, Toth, E.A, MacKerell Jr, A.D, Coop, A, Weber, D.J. | | Deposit date: | 2015-09-03 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.742 Å) | | Cite: | Small Molecule Inhibitors of Ca(2+)-S100B Reveal Two Protein Conformations.
J.Med.Chem., 59, 2016
|
|
6GS2
 
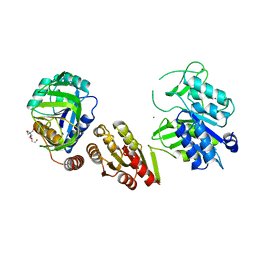 | | Crystal Structure of the GatD/MurT Enzyme Complex from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, SA1707 protein, ... | | Authors: | Muckenfuss, L.M, Noeldeke, E.R, Niemann, V, Zocher, G, Stehle, T. | | Deposit date: | 2018-06-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus.
Sci Rep, 8, 2018
|
|
5AIJ
 
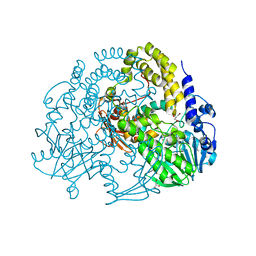 | | P. aeruginosa SdsA hexagonal polymorph | | Descriptor: | ALKYL SULFATASE, GLYCEROL, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
2ATI
 
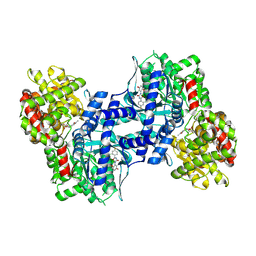 | | Glycogen Phosphorylase Inhibitors | | Descriptor: | Glycogen phosphorylase, liver form, N-(2-CHLORO-4-FLUOROBENZOYL)-N'-(5-HYDROXY-2-METHOXYPHENYL)UREA, ... | | Authors: | Klabunde, T, Wendt, K.U, Kadereit, D, Brachvogel, V, Burger, H.J, Herling, A.W, Oikonomakos, N.G, Schmoll, D, Sarubbi, E, von Roedern, E, Schoenafinger, K, Defossa, E. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Acyl ureas as human liver glycogen phosphorylase inhibitors for the treatment of type 2 diabetes.
J.Med.Chem., 48, 2005
|
|
6H65
 
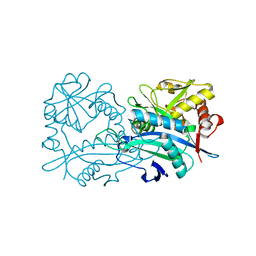 | | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Timofeev, V.I, Bezsudnova, E.Y, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum
To Be Published
|
|
6UB0
 
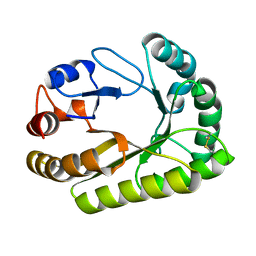 | | Crystal structure of a GH128 (subgroup III) curdlan-specific exo-beta-1,3-glucanase from Blastomyces gilchristii (BgGH128_III) in complex with laminaribiose at -2 and -1 subsites | | Descriptor: | Glyco_hydro_cc domain-containing protein, beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Costa, P.A.C.R, Santos, C.R, Domingues, M.N, Lima, E.A, Mandelli, F, Murakami, M.T. | | Deposit date: | 2019-09-11 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural insights into beta-1,3-glucan cleavage by a glycoside hydrolase family.
Nat.Chem.Biol., 16, 2020
|
|
6UCU
 
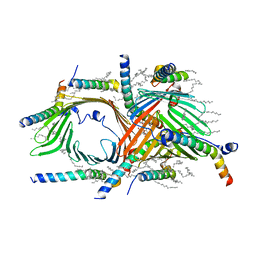 | | Cryo-EM structure of the mitochondrial TOM complex from yeast (dimer) | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, ... | | Authors: | Park, E, Tucker, K. | | Deposit date: | 2019-09-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5I74
 
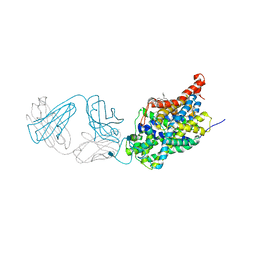 | | X-ray structure of the ts3 human serotonin transporter complexed with Br-citalopram at the central site | | Descriptor: | (1S)-1-(4-bromophenyl)-1-[3-(dimethylamino)propyl]-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.395 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
2WCF
 
 | | calcium-free (apo) S100A12 | | Descriptor: | PROTEIN S100-A12, SODIUM ION | | Authors: | Moroz, O.V, Blagova, E.V, Wilkinson, A.J, Wilson, K.S, Bronstein, I.B. | | Deposit date: | 2009-03-11 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | The Crystal Structures of Human S100A12 in Apo Form and in Complex with Zinc: New Insights Into S100A12 Oligomerisation.
J.Mol.Biol., 391, 2009
|
|
5WGY
 
 | | Crystal Structure of MalA' C112S/C128S, malbrancheamide B complex | | Descriptor: | (5aS,12aS,13aS)-9-chloro-12,12-dimethyl-2,3,11,12,12a,13-hexahydro-1H,5H,6H-5a,13a-(epiminomethano)indolizino[7,6-b]carbazol-14-one, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Fraley, A.E, Smith, J.L. | | Deposit date: | 2017-07-14 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Function and Structure of MalA/MalA', Iterative Halogenases for Late-Stage C-H Functionalization of Indole Alkaloids.
J. Am. Chem. Soc., 139, 2017
|
|
5BW9
 
 | | Crystal Structure of Yeast V1-ATPase in the Autoinhibited Form | | Descriptor: | V-type proton ATPase catalytic subunit A, V-type proton ATPase subunit B, V-type proton ATPase subunit D, ... | | Authors: | Oot, R.A, Kane, P.M, Berry, E.A, Wilkens, S. | | Deposit date: | 2015-06-06 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | Crystal structure of yeast V1-ATPase in the autoinhibited state.
Embo J., 35, 2016
|
|
6TZH
 
 | | ADC-7 in complex with boronic acid transition state inhibitor S06015 | | Descriptor: | Beta-lactamase, GLYCINE, PHOSPHATE ION, ... | | Authors: | Fish, E.R, Powers, R.A, Wallar, B.J. | | Deposit date: | 2019-08-12 | | Release date: | 2020-06-24 | | Last modified: | 2020-07-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | 1,2,3-Triazolylmethaneboronate: A Structure Activity Relationship Study of a Class of beta-Lactamase Inhibitors againstAcinetobacter baumanniiCephalosporinase.
Acs Infect Dis., 6, 2020
|
|
5UIP
 
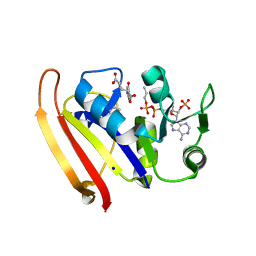 | | structure of DHFR with bound DAP, p-ABG and NADP | | Descriptor: | Dihydrofolate reductase, N-(4-aminobenzene-1-carbonyl)-L-glutamic acid, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2017-01-14 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural Basis for Biguanide Activity.
Biochemistry, 56, 2017
|
|
5I71
 
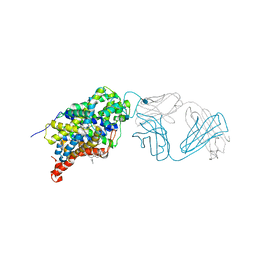 | | X-ray structure of the ts3 human serotonin transporter complexed with s-citalopram at the central site | | Descriptor: | (1S)-1-[3-(dimethylamino)propyl]-1-(4-fluorophenyl)-1,3-dihydro-2-benzofuran-5-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8B6 antibody, ... | | Authors: | Coleman, J.A, Green, E.M, Gouaux, E. | | Deposit date: | 2016-02-16 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | X-ray structures and mechanism of the human serotonin transporter.
Nature, 532, 2016
|
|
6D74
 
 | |
4GX9
 
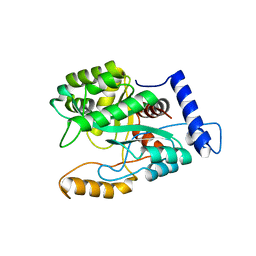 | | Crystal structure of a DNA polymerase III alpha-epsilon chimera | | Descriptor: | DNA polymerase III subunit epsilon,DNA polymerase III subunit alpha | | Authors: | Li, N, Horan, N, Xu, Z.-Q, Jacques, D, Dixon, N.E, Oakley, A.J. | | Deposit date: | 2012-09-04 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proofreading exonuclease on a tether: the complex between the E. coli DNA polymerase III subunits alpha, {varepsilon}, theta and beta reveals a highly flexible arrangement of the proofreading domain
Nucleic Acids Res., 41, 2013
|
|
