7AXM
 
 | | Structure of SARS-CoV-2 Main Protease bound to Pelitinib | | Descriptor: | (2E)-N-{4-[(3-chloro-4-fluorophenyl)amino]-3-cyano-7-ethoxyquinolin-6-yl}-4-(dimethylamino)but-2-enamide, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | Authors: | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
8S7O
 
 | | M. tuberculosis gyrase holocomplex with 150 bp DNA and BDM71403 | | Descriptor: | 6-[[2-[1-(6-methoxy-1,5-naphthyridin-4-yl)-1,2,3-triazol-4-yl]ethylamino]methyl]-4H-1,4-benzothiazin-3-one, DNA (5'-D(*CP*CP*GP*GP*AP*AP*GP*GP*GP*GP*TP*AP*AP*TP*AP*CP*T)-3'), DNA gyrase subunit A, ... | | Authors: | Gedeon, A, Yab, E, Dinut, A, Sadowski, E, Capton, E, Dreneau, A, Gioia, B, Piveteau, C, Djaout, K, Lecat, E, Wehenkel, A.M, Gubellini, F, Mechaly, A, Alzari, P.M, Deprez, B, Baulard, A, Aubry, A, Willand, N, Petrella, S. | | Deposit date: | 2024-03-04 | | Release date: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | M. tuberculosis gyrase holocomplex with 150 bp DNA and BDM71403
To Be Published
|
|
8PMV
 
 | | transcription factor BARHL2 bound to TAAGC DNA sequence | | Descriptor: | BarH-like 2 homeobox protein, DNA | | Authors: | Morgunova, E, Popov, A, Yin, Y, Taipale, J. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-10 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interfacial water confers transcription factors with dinucleotide specificity.
Nat.Struct.Mol.Biol., 32, 2025
|
|
7QJR
 
 | | Crystal structure of cutinase 1 from Thermobifida fusca DSM44342 (703) | | Descriptor: | Cutinase 1, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJP
 
 | | Crystal structure of a cutinase enzyme from Saccharopolyspora flava (611) | | Descriptor: | Cutinase, TETRAETHYLENE GLYCOL | | Authors: | Zahn, M, Avilan, L, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2021-12-17 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.561 Å) | | Cite: | Sourcing thermotolerant poly(ethylene terephthalate) hydrolase scaffolds from natural diversity
Nat Commun, 13, 2022
|
|
7QJO
 
 | |
6QA6
 
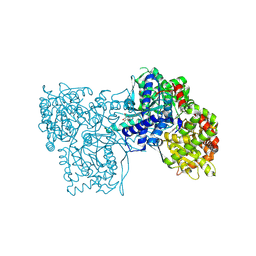 | | Glycogen Phosphorylase b in complex with 30 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-2-naphthalen-2-yl-8,9,10-tris(oxidanyl)-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
7QJM
 
 | |
6PWQ
 
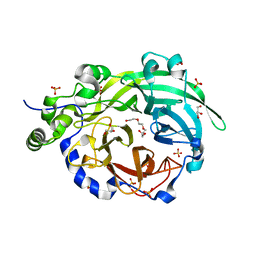 | | Crystal structure of Levansucrase from Bacillus subtilis mutant S164A at 2.6 A | | Descriptor: | CALCIUM ION, GLYCEROL, Glycoside hydrolase family 68 protein, ... | | Authors: | Diaz-Vilchis, A, Rodriguez-Alegria, M.E, Ortiz-Soto, M.E, Rudino-Pinera, E, Lopez-Munguia, A. | | Deposit date: | 2019-07-23 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Implications of the mutation S164A on Bacillus subtilis levansucrase product specificity and insights into protein interactions acting upon levan synthesis.
Int.J.Biol.Macromol., 161, 2020
|
|
8OS6
 
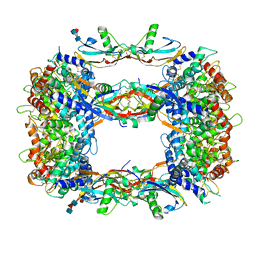 | | Structure of a GFRA1/GDNF LICAM complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Houghton, F.M, Adams, S.E, Briggs, D.C, McDonald, N.Q. | | Deposit date: | 2023-04-18 | | Release date: | 2023-11-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Architecture and regulation of a GDNF-GFR alpha 1 synaptic adhesion assembly.
Nat Commun, 14, 2023
|
|
4YO3
 
 | | Enteroaggregative Escherichia Coli TssA N-terminal fragment | | Descriptor: | TssA | | Authors: | Durand, E, Zoued, A, Spinelli, S, Douzi, B, Brunet, Y.R, Bebeacua, C, Legrand, P, Journet, L, Mignot, T, Cambillau, C, Cascales, E. | | Deposit date: | 2015-03-11 | | Release date: | 2016-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.37 Å) | | Cite: | Priming and polymerization of a bacterial contractile tail structure.
Nature, 531, 2016
|
|
8CDM
 
 | | Plasmodium falciparum Myosin A full-length, post-rigor state complexed to the inhibitor KNX-002 | | Descriptor: | 1,2-ETHANEDIOL, 1-(4-methoxyphenyl)-~{N}-[(3-thiophen-2-yl-1~{H}-pyrazol-4-yl)methyl]cyclopropan-1-amine, Myosin A tail domain interacting protein, ... | | Authors: | Moussaoui, D, Robblee, J.P, Robert-Paganin, J, Auguin, D, Fisher, F, Fagnant, P.M, MacFarlane, J.E, Mueller-Dieckmann, C, Baum, J, Trybus, K.M, Houdusse, A. | | Deposit date: | 2023-01-31 | | Release date: | 2024-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Mechanism of small molecule inhibition of Plasmodium falciparum myosin A informs antimalarial drug design.
Nat Commun, 14, 2023
|
|
6YE1
 
 | | Human Ecto-5'-nucleotidase (CD73) in complex with the AMPCP derivative A894 (compound 2n in publication) in the closed form (crystal form IV) | | Descriptor: | 5'-nucleotidase, ZINC ION, [(2~{R},3~{S},4~{R},5~{R})-5-[2-chloranyl-6-(cyclopentylamino)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxymethylphosphonic acid | | Authors: | Scaletti, E, Strater, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Discovery of Potent and Selective Methylenephosphonic Acid CD73 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7R6X
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2E12 Fab, S309 Fab, and S304 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Monoclonal antibody S2E12 Fab heavy chain, ... | | Authors: | Snell, G, Czudnochowski, N, Croll, T.I, Nix, J.C, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7R6W
 
 | | SARS-CoV-2 spike receptor-binding domain (RBD) in complex with S2X35 Fab and S309 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Snell, G, Czudnochowski, N, Hernandez, P, Nix, J.C, Croll, T.I, Corti, D, Cameroni, E, Pinto, D, Beltramello, M. | | Deposit date: | 2021-06-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape.
Nature, 597, 2021
|
|
7OP0
 
 | | Crystal structure of complement C5 in complex with chemically synthesized K92 knob domain. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C5 alpha chain, ... | | Authors: | Macpherson, A, van der Elsen, J.M.H, Schulze, M.E, Birtley, J.R. | | Deposit date: | 2021-05-28 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The Chemical Synthesis of Knob Domain Antibody Fragments.
Acs Chem.Biol., 16, 2021
|
|
8BZH
 
 | | FC-NAc stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
7P0X
 
 | | Crystal structure of Thioredoxin reductase from Brugia Malayi | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Cheng, Q, Arner, E.S.J, Williams, D.L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
7OBO
 
 | | GSTF1 from Alopecurus myosuroides | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-3-[(7-nitro-2,1,3-benzoxadiazol-4-yl)sulfanyl]-1-oxidanylidene-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, Glutathione transferase | | Authors: | Pohl, E, Eno, R.F.M, Freitag-Pohl, S. | | Deposit date: | 2021-04-23 | | Release date: | 2022-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
7ODM
 
 | | AmGSTF1 Y118S variant | | Descriptor: | Glutathione transferase, [(2~{S})-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-sulfanyl-propan-2-yl]amino]-1-oxidanyl-1,5-bis(oxidanylidene)pentan-2-yl]azanium | | Authors: | Pohl, E, Eno, R.F.M. | | Deposit date: | 2021-04-30 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Flavonoid-based inhibitors of the Phi-class glutathione transferase from black-grass to combat multiple herbicide resistance.
Org.Biomol.Chem., 19, 2021
|
|
8BZE
 
 | | FC-J stabilizer for ERa and 14-3-3 | | Descriptor: | (2~{S},3~{R},4~{S},5~{S},6~{R})-2-[[(1~{E},3~{R},4~{S},8~{R},9~{R},10~{R},11~{S},14~{S})-14-(methoxymethyl)-3,10-dimethyl-4,9-bis(oxidanyl)-6-propan-2-yl-8-tricyclo[9.3.0.0^{3,7}]tetradeca-1,6-dienyl]oxy]-6-(2-methylbut-3-en-2-yloxymethyl)oxane-3,4,5-triol, 14-3-3 protein sigma, ERalpha peptide, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a new potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
7RA4
 
 | | Crystal structure of Neisseria gonorrhoeae serine acetyltransferase (CysE) in complex with serine | | Descriptor: | SERINE, Serine acetyltransferase | | Authors: | Hicks, J.L, Oldham, K.E, Prentice, E.J, Summers, E.L. | | Deposit date: | 2021-06-30 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Serine acetyltransferase from Neisseria gonorrhoeae; structural and biochemical basis of inhibition.
Biochem.J., 479, 2022
|
|
8BZT
 
 | | FC-NAg stabilizer of 14-3-3 and ERalpha | | Descriptor: | 14-3-3 protein sigma, ERalpha peptide, MAGNESIUM ION, ... | | Authors: | Visser, E.J, Ottmann, C. | | Deposit date: | 2022-12-15 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stabilization of the Estrogen receptor alpha - 14-3-3 interaction as a potential intervention strategy for endocrine resistance in breast cancer
To Be Published
|
|
8OLD
 
 | | Crystal structure of Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Archaeoglobus fulgidus AfAgo-N protein representing N-L1-L2 domains, CACODYLATE ION, ... | | Authors: | Manakova, E.N, Zaremba, M, Grazulis, S. | | Deposit date: | 2023-03-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The missing part: the Archaeoglobus fulgidus Argonaute forms a functional heterodimer with an N-L1-L2 domain protein.
Nucleic Acids Res., 52, 2024
|
|
6GFI
 
 | | Structure of Human Mesotrypsin in complex with APPI variant T11V/M17R/I18F/F34V | | Descriptor: | 1,2-ETHANEDIOL, Amyloid-beta A4 protein, PRSS3 protein | | Authors: | Shahar, A, Cohen, I, Radisky, E, Papo, N, Naftaly, S. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mapping protein selectivity landscapes using multi-target selective screening and next-generation sequencing of combinatorial libraries.
Nat Commun, 9, 2018
|
|
