1W4T
 
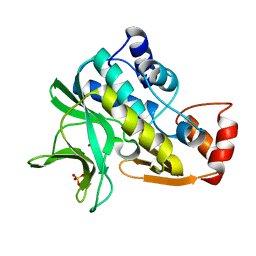 | | X-ray crystallographic structure of Pseudomonas aeruginosa arylamine N-acetyltransferase | | Descriptor: | Arylamine N-acetyltransferase, SULFATE ION | | Authors: | Westwood, I.M, Holton, S.J, Rodrigues-Lima, F, Dupret, J.-M, Bhakta, S, Noble, M.E, Sim, E. | | Deposit date: | 2004-07-29 | | Release date: | 2005-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Expression, purification, characterization and structure of Pseudomonas aeruginosa arylamine N-acetyltransferase.
Biochem. J., 385, 2005
|
|
1MDX
 
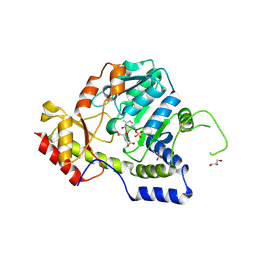 | | Crystal structure of ArnB transferase with pyridoxal 5' phosphate | | Descriptor: | 2-OXOGLUTARIC ACID, ArnB aminotransferase, GLYCEROL, ... | | Authors: | Noland, B.W, Newman, J.M, Hendle, J, Badger, J, Christopher, J.A, Tresser, J, Buchanan, M.D, Wright, T.A, Rutter, M.E, Sanderson, W.E, Muller-Dieckmann, H.-J, Gajiwala, K.S, Sauder, J.M, Buchanan, S.G. | | Deposit date: | 2002-08-07 | | Release date: | 2002-12-11 | | Last modified: | 2018-12-26 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural studies of Salmonella typhimurium ArnB (PmrH) aminotransferase: A 4-amino-4-deoxy-L-arabinose lipopolysaccharide modifying enzyme
Structure, 10, 2002
|
|
3CDE
 
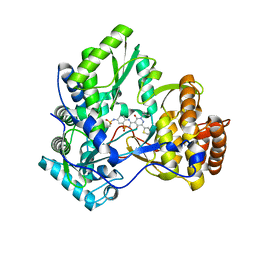 | | Crystal structure of HCV NS5B polymerase with a novel Pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-02-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 3: Further optimization of the 2-, 6-, and 7'-substituents and initial pharmacokinetic assessments.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7OJL
 
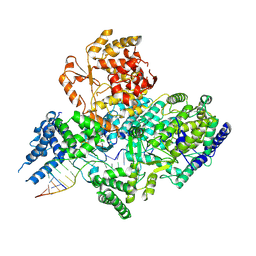 | | Lassa virus L protein in a pre-initiation conformation [PREINITIATION] | | Descriptor: | 3' RNA, 5' RNA, MANGANESE (II) ION, ... | | Authors: | Kouba, T, Vogel, D, Thorkelsson, S, Quemin, E, Williams, H.M, Milewski, M, Busch, C, Gunther, S, Grunewald, K, Rosenthal, M, Cusack, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-12-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational changes in Lassa virus L protein associated with promoter binding and RNA synthesis activity.
Nat Commun, 12, 2021
|
|
1MK5
 
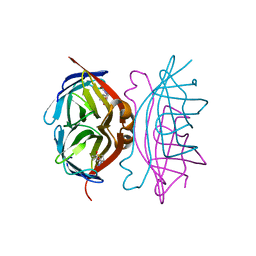 | | Wildtype Core-Streptavidin with Biotin at 1.4A. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Green, N.M, Stenkamp, R.E, Stayton, P.S. | | Deposit date: | 2002-08-28 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
5LRW
 
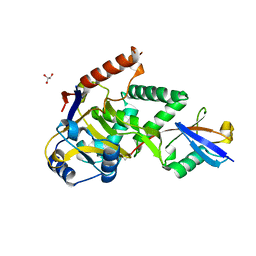 | | Structure of Cezanne/OTUD7B OTU domain bound to ubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, Polyubiquitin-B | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
4MF3
 
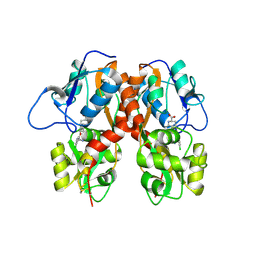 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1MD2
 
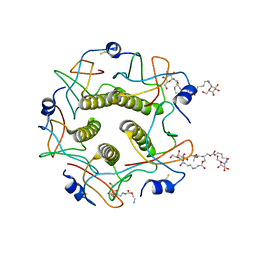 | | CHOLERA TOXIN B-PENTAMER WITH DECAVALENT LIGAND BMSC-0013 | | Descriptor: | 3-ETHYLAMINO-4-METHYLAMINO-CYCLOBUTANE-1,2-DIONE, CHOLERA TOXIN B SUBUNIT, CYANIDE ION, ... | | Authors: | Zhang, Z, Merritt, E.A, Ahn, M, Roach, C, Hol, W.G.J, Fan, E. | | Deposit date: | 2002-08-06 | | Release date: | 2002-12-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Solution and crystallographic studies of branched multivalent ligands that inhibit the receptor-binding of cholera toxin.
J.Am.Chem.Soc., 124, 2002
|
|
1MEP
 
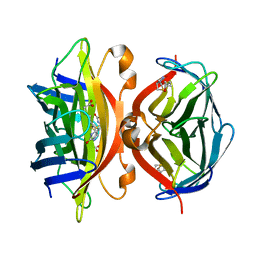 | | Crystal Structure of Streptavidin Double Mutant S45A/D128A with Biotin: Cooperative Hydrogen-Bond Interactions in the Streptavidin-Biotin System. | | Descriptor: | BIOTIN, Streptavidin | | Authors: | Hyre, D.E, Le Trong, I, Merritt, E.A, Stenkamp, R.E, Green, N.M, Stayton, P.S. | | Deposit date: | 2002-08-08 | | Release date: | 2003-09-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Cooperative hydrogen bond interactions in the streptavidin-biotin system
Protein Sci., 15, 2006
|
|
1M5W
 
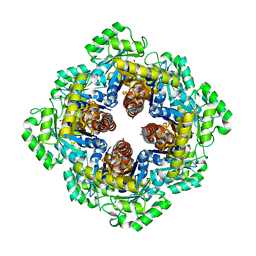 | | 1.96 A Crystal Structure of Pyridoxine 5'-Phosphate Synthase in Complex with 1-deoxy-D-xylulose phosphate | | Descriptor: | 1-DEOXY-D-XYLULOSE-5-PHOSPHATE, PHOSPHATE ION, Pyridoxal phosphate biosynthetic protein pdxJ | | Authors: | Yeh, J.I, Du, S, Pohl, E, Cane, D.E. | | Deposit date: | 2002-07-10 | | Release date: | 2003-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multistate Binding in Pyridoxine 5'-Phosphate Synthase: 1.96 A Crystal Structure in
Complex with 1-deoxy-D-xylulose phosphate
Biochemistry, 41, 2002
|
|
4W8J
 
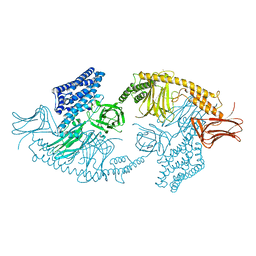 | | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals | | Descriptor: | CALCIUM ION, POTASSIUM ION, Pesticidal crystal protein cry1Ac | | Authors: | Evdokimov, A.G, Moshiri, F, Sturman, E.J, Rydel, T.J, Zheng, M, Seale, J.W, Franklin, S. | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of the full-length insecticidal protein Cry1Ac reveals intriguing details of toxin packaging into in vivo formed crystals.
Protein Sci., 23, 2014
|
|
2QH7
 
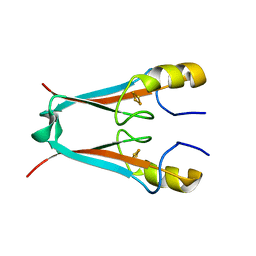 | | MitoNEET is a uniquely folded 2Fe-2S outer mitochondrial membrane protein stabilized by pioglitazone | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Zinc finger CDGSH-type domain 1 | | Authors: | Paddock, M.L, Wiley, S.E, Axelrod, H.L, Cohen, A.E, Roy, M, Abresch, E.C, Capraro, D, Murphy, A.N, Nechushtai, R, Dixon, J.E, Jennings, P.A. | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | MitoNEET is a uniquely folded 2Fe 2S outer mitochondrial membrane protein stabilized by pioglitazone.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4W8W
 
 | |
4WCY
 
 | | Fab fragment of mouse AZ130 monoclonal antibody | | Descriptor: | AZ130 Heavy chain, AZ130 Light chain, GLYCEROL, ... | | Authors: | Fallecker, C, Tarbouriech, N, Habib, M, Crepin, T, Drouet, E. | | Deposit date: | 2014-09-05 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Fab fragment of mouse AZ130 monoclonal antibody
To Be Published
|
|
5LG3
 
 | | X-ray structure of a pentameric ligand gated ion channel from Erwinia chrysanthemi (ELIC) in complex with chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Nys, M, Wijckmans, E, Farinha, A, Brams, M, Spurny, R, Ulens, C. | | Deposit date: | 2016-07-05 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.567 Å) | | Cite: | Allosteric binding site in a Cys-loop receptor ligand-binding domain unveiled in the crystal structure of ELIC in complex with chlorpromazine.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4UXU
 
 | | Crystal Structure of the Extracellular Domain of the Human Alpha9 Nicotinic Acetylcholine Receptor In Complex with Methyllycaconitine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Zouridakis, M, Giastas, P, Zarkadas, E, Tzartos, S.J. | | Deposit date: | 2014-08-27 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Nat.Struct.Mol.Biol., 21, 2014
|
|
2QQD
 
 | | N47A mutant of Pyruvoyl-dependent Arginine Decarboxylase from Methanococcus jannashii | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AGMATINE, PYRUVIC ACID, ... | | Authors: | Ealick, S.E, Soriano, E.S. | | Deposit date: | 2007-07-26 | | Release date: | 2008-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the N47A and E109Q mutant proteins of pyruvoyl-dependent arginine decarboxylase from Methanococcus jannaschii.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
4UUP
 
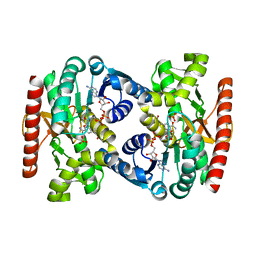 | | Reconstructed ancestral trichomonad malate dehydrogenase in complex with NADH, SO4, and PO4 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, PHOSPHATE ION, ... | | Authors: | Steindel, P.A, Chen, E.H, Theobald, D.L. | | Deposit date: | 2014-07-29 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Gradual Neofunctionalization in the Convergent Evolution of Trichomonad Lactate and Malate Dehydrogenases.
Protein Sci., 25, 2016
|
|
4UYZ
 
 | |
4V2K
 
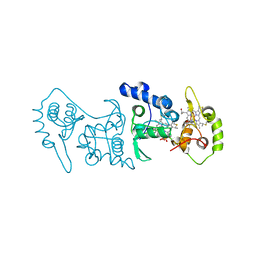 | | Crystal structure of the thiosulfate dehydrogenase TsdA in complex with thiosulfate | | Descriptor: | HEME C, THIOSULFATE, THIOSULFATE DEHYDROGENASE | | Authors: | Grabarczyk, D.B, Chappell, P.E, Eisel, B, Johnson, S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-10-10 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Mechanism of Thiosulfate Oxidation in the Soxa Family of Cysteine-Ligated Cytochromes
J.Biol.Chem., 290, 2015
|
|
4V6R
 
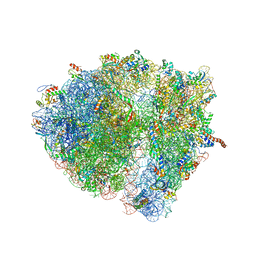 | | Structural characterization of mRNA-tRNA translocation intermediates (class 6 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V9Q
 
 | | Crystal Structure of Blasticidin S Bound to Thermus Thermophilus 70S Ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Svidritskiy, E, Ling, C, Ermolenko, D.N, Korostelev, A.A. | | Deposit date: | 2013-06-12 | | Release date: | 2014-07-09 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Blasticidin S inhibits translation by trapping deformed tRNA on the ribosome.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4W80
 
 | |
4UR9
 
 | | Structure of ligand bound glycosylhydrolase | | Descriptor: | 4-ethoxyquinazoline, CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, ... | | Authors: | Darby, J.F, Landstroem, J, Roth, C, He, Y, Schultz, M, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2014-06-27 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of Selective Small-Molecule Activators of a Bacterial Glycoside Hydrolase.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4UQZ
 
 | | Coevolution of the ATPase ClpV, the TssB-TssC Sheath and the Accessory HsiE Protein Distinguishes Two Type VI Secretion Classes | | Descriptor: | ACETATE ION, HSIB1, HSIE1 | | Authors: | Forster, A, Planamente, S, Manoli, E, Lossi, N.S, Freemont, P.S, Filloux, A. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Coevolution of the ATPase Clpv, the Sheath Proteins Tssb and Tssc and the Accessory Protein Tagj/Hsie1 Distinguishes Type Vi Secretion Classes.
RNA, 20, 2014
|
|
