7F4B
 
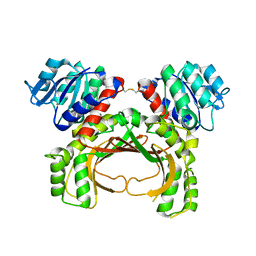 | | The crystal structure of the immature apo-enzyme of homoserine dehydrogenase from the hyperthermophilic archaeon Sulfurisphaera tokodaii. | | Descriptor: | MAGNESIUM ION, homoserine dehydrogenase | | Authors: | Kurihara, E, Kubota, T, Watanabe, K, Ogata, K, Kaneko, R, Oshima, T, Yoshimune, K, Goto, M. | | Deposit date: | 2021-06-18 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Conformational changes in the catalytic region are responsible for heat-induced activation of hyperthermophilic homoserine dehydrogenase.
Commun Biol, 5, 2022
|
|
7F4X
 
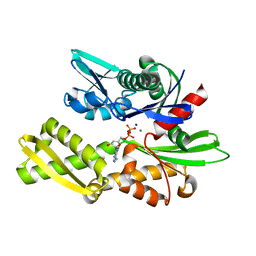 | | Joint neutron and X-ray crystal structure of the nucleotide-binding domain of Hsp72 in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock 70 kDa protein 1B, MAGNESIUM ION, ... | | Authors: | Yokoyama, T, Ostermann, A, Schrader, T.E. | | Deposit date: | 2021-06-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron crystallographic analysis of the nucleotide-binding domain of Hsp72 in complex with ADP.
Iucrj, 9, 2022
|
|
8A2X
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP(S)-2'F,2'dAMP(S)) | | Descriptor: | 9-[(1~{R},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]purin-6-amine, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-06-06 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP(S)-2'F,2'dAMP(S))
To Be Published
|
|
8BLR
 
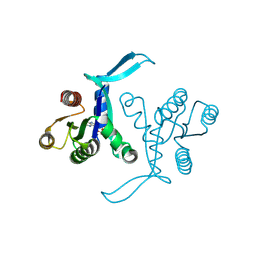 | | G13D mutant of KRAS4b (2-169) bound to GDP with the switch-I in fully open conformation | | Descriptor: | GTPase KRas, N-terminally processed, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Moche, M, Jungholm, O, Strandback, E, Ampah-Korsah, H, Nyman, T, Orwar, O. | | Deposit date: | 2022-11-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel druggable space in human KRAS G13D discovered using structural bioinformatics and a P-loop targeting monoclonal antibody.
Sci Rep, 14, 2024
|
|
6L27
 
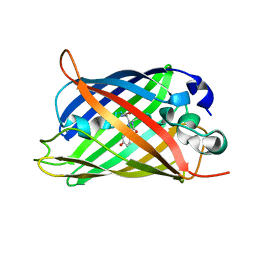 | | X-ray crystal structure of the mutant green fluorescent protein | | Descriptor: | Green fluorescent protein | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.77 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
8BFM
 
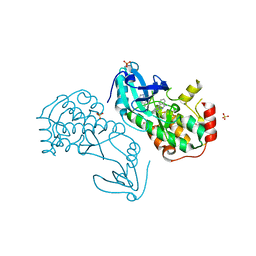 | | Crystal structure of human calmodulin-dependent protein kinase 1D (CAMK1D) in complex with FZ331 | | Descriptor: | Calcium/calmodulin-dependent protein kinase type 1D, SULFATE ION, pyrazolo[5,1-a]phthalazin-6-amine | | Authors: | Kraemer, A, Zhu, W.F, Hernandez-Olmos, V, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human WDR5 and pVHL:ElonginC:ElonginB bound to PROTAC with PEG linker (conformation #2)
To Be Published
|
|
6L26
 
 | | Neutron crystal structure of the mutant green fluorescent protein (EGFP) | | Descriptor: | Green fluorescent protein, trideuteriooxidanium | | Authors: | Adachi, M, Shimizu, R, Shibazaki, C, Kagotani, Y, Ostermann, A, Schrader, T.E. | | Deposit date: | 2019-10-02 | | Release date: | 2020-04-08 | | Last modified: | 2025-01-01 | | Method: | NEUTRON DIFFRACTION (1.444 Å) | | Cite: | Direct Observation of the Protonation States in the Mutant Green Fluorescent Protein.
J Phys Chem Lett, 11, 2020
|
|
6L62
 
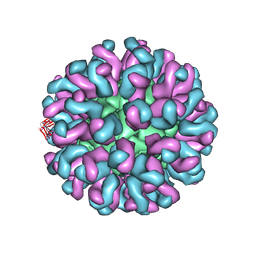 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
6TN7
 
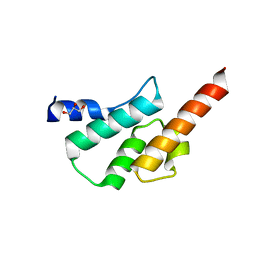 | | Crystal structure of the human Arc C-lobe | | Descriptor: | Activity-regulated cytoskeleton-associated protein, GLYCEROL | | Authors: | Hallin, E.I, Bramham, C.R, Kursula, P. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural properties and peptide ligand binding of the capsid homology domains of human Arc.
Biochem Biophys Rep, 26, 2021
|
|
8BFI
 
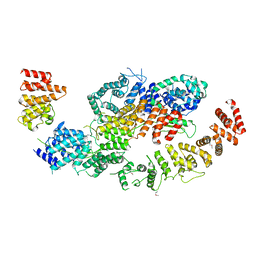 | | human CNOT1-CNOT10-CNOT11 module | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BFH
 
 | | CNOT11 | | Descriptor: | CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
8BDZ
 
 | | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=4 topology | | Descriptor: | Core protein,Matrix protein 2,External core antigen | | Authors: | Egorov, V.V, Shvetsov, A.V, Pichkur, E.B, Shaldzhyan, A.A, Zabrodskaya, Y.A, Vinogradova, D.S, Nekrasov, P.A, Gorshkov, A.N, Garmay, Y.P, Kovaleva, A.A, Stepanova, L.A, Tsybalova, L.M, Shtam, T.A, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2022-10-20 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure.
Biophys.Chem., 293, 2022
|
|
6TRU
 
 | |
8BER
 
 | | Hepatitis B virus core antigen (HBc) with the insertion of four external domains of the influenza A M2 protein (HBc/4M2e) with T=3 topology | | Descriptor: | Core protein,Matrix protein 2,External core antigen | | Authors: | Egorov, V.V, Shvetsov, A.V, Pichkur, E.B, Shaldzhyan, A.A, Zabrodskaya, Y.A, Vinogradova, D.S, Nekrasov, P.A, Gorshkov, A.N, Garmay, Y.P, Kovaleva, A.A, Stepanova, L.A, Tsybalova, L.M, Shtam, T.A, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2022-10-21 | | Release date: | 2022-12-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inside and outside of virus-like particles HBc and HBc/4M2e: A comprehensive study of the structure.
Biophys.Chem., 293, 2022
|
|
6TJS
 
 | |
6TK8
 
 | |
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
8AZB
 
 | |
7FQM
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000619a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-({[(thiophen-2-yl)methyl]amino}methyl)phenol, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRE
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B after initial refinement with no ligand modeled | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-20 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FQT
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOMB000293a | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRP
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with XST00000245b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-(2-methyl-1,3-thiazol-4-yl)thiophene-2-carboxylic acid, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRU
 
 | | PanDDA analysis group deposition of ground-state model of PTP1B, using pre-clustering, cluster 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FQX
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000601a | | Descriptor: | 1-(3,4-dimethoxyphenyl)methanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FQV
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with XST00000847b | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N,N-dimethylpyridin-4-amine, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
