7KKW
 
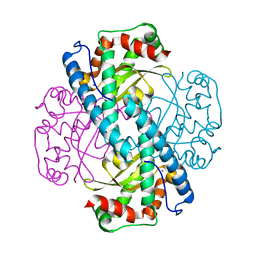 | | Neutron structure of Reduced Human MnSOD | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial, ... | | Authors: | Azadmanesh, J, Lutz, W.E, Coates, L, Weiss, K.L, Borgstahl, G.E.O. | | Deposit date: | 2020-10-28 | | Release date: | 2021-04-21 | | Last modified: | 2024-04-10 | | Method: | NEUTRON DIFFRACTION (2.3 Å) | | Cite: | Direct detection of coupled proton and electron transfers in human manganese superoxide dismutase.
Nat Commun, 12, 2021
|
|
7Z18
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z17
 
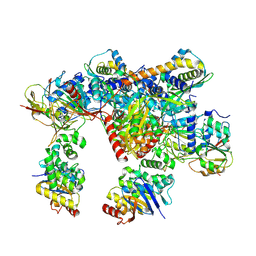 | | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
4Z1R
 
 | | Crystal structure of collagen-like peptide at 1.27 Angstrom resolution | | Descriptor: | Collagen-like peptide | | Authors: | Plonska-Brzezinska, M.E, Czyrko, J, Brus, D.M, Imierska, M, Brzezinski, K. | | Deposit date: | 2015-03-27 | | Release date: | 2015-11-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Triple helical collagen-like peptide interactions with selected polyphenolic compounds.
Rsc Adv, 5, 2015
|
|
7KRP
 
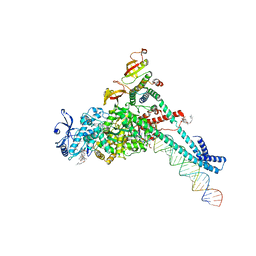 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPSO, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7Z16
 
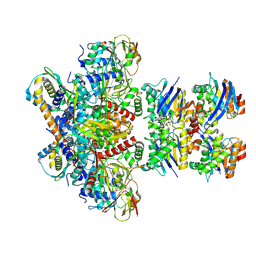 | | E. coli C-P lyase bound to PhnK/PhnL dual ABC dimer with AMPPNP and PhnK E171Q mutation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnI, ... | | Authors: | Amstrup, S.K, Sofus, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
6DTK
 
 | | Heterodimers of FALS mutant SOD enzyme | | Descriptor: | COPPER (II) ION, MALONATE ION, Superoxide dismutase C111S/D83S-C111S HETERODIMER, ... | | Authors: | Streltsov, V.A, Nuttall, S.D, Ganio, K.E, Roberts, B. | | Deposit date: | 2018-06-17 | | Release date: | 2019-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of heterodimers of FALS mutant SOD enzyme
To Be Published
|
|
7KRO
 
 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - nsp13(2)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7Z15
 
 | | E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.93 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
6T3J
 
 | | Dual Epitope Targeting by Anti-DR5 Antibodies | | Descriptor: | IgG1-hDR5-01-Heavy Chain, IgG1-hDR5-01-Light Chain, IgG1-hDR5-05-Heavy Chain, ... | | Authors: | Tauchert, M.J, Augustin, M, Krapp, S, Overdijk, M.B, Breij, E.C.W, Hibbert, R.G. | | Deposit date: | 2019-10-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Dual Epitope Targeting and Enhanced Hexamerization by DR5 Antibodies as a Novel Approach to Induce Potent Antitumor Activity Through DR5 Agonism.
Mol.Cancer Ther., 19, 2020
|
|
5DXM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol | | Descriptor: | 3-[(E)-(1s,5s)-bicyclo[3.3.1]non-9-ylidene(4-hydroxyphenyl)methyl]phenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
7YZN
 
 | | Structure of C-terminally truncated aIF5B from Pyrococcus abyssi complexed with GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Probable translation initiation factor IF-2, ... | | Authors: | Bourgeois, G, Schmitt, E, Mechulam, Y, Coureux, P.D, Kazan, R. | | Deposit date: | 2022-02-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Role of aIF5B in archaeal translation initiation.
Nucleic Acids Res., 50, 2022
|
|
1B72
 
 | | PBX1, HOMEOBOX PROTEIN HOX-B1/DNA TERNARY COMPLEX | | Descriptor: | DNA (5'-D(*AP*CP*TP*CP*TP*AP*TP*GP*AP*TP*TP*GP*AP*TP*CP*GP*GP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*GP*CP*CP*GP*AP*TP*CP*AP*AP*TP*CP*AP*TP*AP*GP*AP*G)-3'), PROTEIN (HOMEOBOX PROTEIN HOX-B1), ... | | Authors: | Piper, D.E, Batchelor, A.H, Chang, C.-P, Cleary, M.L, Wolberger, C. | | Deposit date: | 1999-01-27 | | Release date: | 1999-02-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation.
Cell(Cambridge,Mass.), 96, 1999
|
|
4Z6U
 
 | | Structure of H200E variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-nitrocatechol at 1.48 Ang resolution | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
3KS8
 
 | | Crystal structure of Reston ebolavirus VP35 RNA binding domain in complex with 18bp dsRNA | | Descriptor: | 5'-R(*AP*GP*AP*AP*GP*GP*AP*GP*GP*GP*AP*GP*GP*GP*AP*GP*GP*A)-3', 5'-R(*UP*CP*CP*UP*CP*CP*CP*UP*CP*CP*CP*UP*CP*CP*UP*UP*CP*U)-3', Polymerase cofactor VP35 | | Authors: | Kimberlin, C.R, Bornholdt, Z.A, Li, S, Woods, V.L, Macrae, I.J, Saphire, E.O. | | Deposit date: | 2009-11-20 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression.
Proc.Natl.Acad.Sci.USA, 107, 2009
|
|
6EU3
 
 | | Apo RNA Polymerase III - closed conformation (cPOL3) | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Abascal-Palacios, G, Ramsay, E.P, Beuron, F, Morris, E, Vannini, A. | | Deposit date: | 2017-10-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase III transcription initiation.
Nature, 553, 2018
|
|
5KE3
 
 | | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a | | Descriptor: | (S)-N-(furan-2-ylmethyl)-1-(1,2,3,4-tetrahydroisoquinoline-3-carbonyl)piperidine-4-carboxamide, BETA-MERCAPTOETHANOL, Histone-lysine N-methyltransferase SETDB1, ... | | Authors: | Dong, A, Iqbal, A, Mader, P, Dobrovetsky, E, Ferreira de Freitas, R, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Schapira, M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-06-09 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SETDB1 Tudor domain in complex with fragment MRT0181a
to be published
|
|
6F2K
 
 | | Crystal structure of Hen Egg-White Lysozyme co-crystallized in presence of 100 mM Tb-Xo4 and 100 mM potassium phosphate monobasic. | | Descriptor: | CHLORIDE ION, Lysozyme C, TERBIUM(III) ION | | Authors: | Engilberge, S, Riobe, F, Di Pietro, S, Girard, E, Dumont, E, Maury, O. | | Deposit date: | 2017-11-24 | | Release date: | 2018-10-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unveiling the Binding Modes of the Crystallophore, a Terbium-based Nucleating and Phasing Molecular Agent for Protein Crystallography.
Chemistry, 24, 2018
|
|
7NM0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(2,6-dihydroxyphenyl)ethan-1-one. | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(oxidanyl)phenyl]ethanone, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4RHV
 
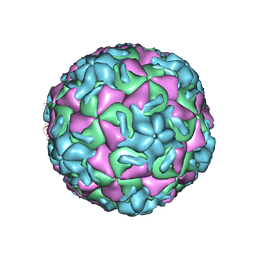 | |
7NLY
 
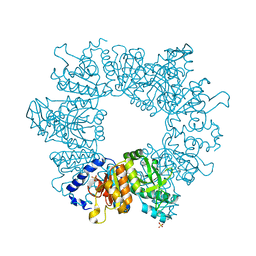 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-Chlorobenzimidazole. | | Descriptor: | 2-chloranyl-1~{H}-benzimidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.246 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4Z8V
 
 | | CRYSTAL STRUCTURE OF AVRRXO1-ORF1:-ORF2 COMPLEX, NATIVE. | | Descriptor: | AvrRxo1-ORF1, AvrRxo1-ORF2, PHOSPHATE ION | | Authors: | Han, Q, Zhou, C, Wu, S, Liu, Y, Yang, Z, Miao, J, Triplett, L, Cheng, Q, Tokuhisa, J, Deblais, L, Robinson, H, Leach, J.E, Li, J, Zhao, B. | | Deposit date: | 2015-04-09 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Xanthomonas AvrRxo1-ORF1, a Type III Effector with a Polynucleotide Kinase Domain, and Its Interactor AvrRxo1-ORF2.
Structure, 23, 2015
|
|
7NNB
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2,8-bis(trifluoromethyl)quinolin-4-ol. | | Descriptor: | 1,2-ETHANEDIOL, 2,8-bis(trifluoromethyl)quinolin-4-ol, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
6EXY
 
 | | Neutron crystal structure of perdeuterated galectin-3C in complex with glycerol | | Descriptor: | GLYCEROL, Galectin-3 | | Authors: | Manzoni, F, Schrader, T.E, Ostermann, A, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-10 | | Release date: | 2018-09-12 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.1 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
1KG5
 
 | | Crystal structure of the K142Q mutant of E.coli MutY (core fragment) | | Descriptor: | A/G-specific adenine glycosylase, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Gilboa, R, Kilshtein, A, Zharkov, D.O, Kycia, J.H, Gerchman, S.E, Grollman, A.P, Shoham, G. | | Deposit date: | 2001-11-26 | | Release date: | 2002-11-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Analysis of the E.coli MutY DNA glycosylase structure and function by site-directed mutagenesis
To be Published
|
|
