1W9R
 
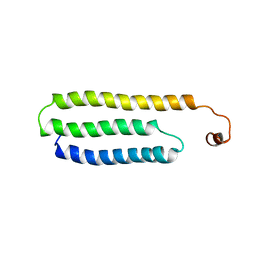 | | Solution Structure of Choline Binding Protein A, Domain R2, the Major Adhesin of Streptococcus pneumoniae | | Descriptor: | CHOLINE BINDING PROTEIN A | | Authors: | Luo, R, Mann, B, Lewis, W.S, Rowe, A, Heath, R, Stewart, M.L, Hamburger, A.E, Bjorkman, P.J, Sivakolundu, S, Lacy, E.R, Tuomanen, E, Kriwacki, R.W. | | Deposit date: | 2004-10-15 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Choline Binding Protein A, the Major Adhesin of Streptococcus Pneumoniae
Embo J., 24, 2005
|
|
1I7M
 
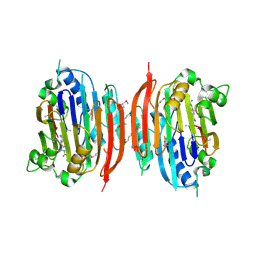 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COMPLEXED WITH 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE | | Descriptor: | 1,4-DIAMINOBUTANE, 4-AMIDINOINDAN-1-ONE-2'-AMIDINOHYDRAZONE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-09 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
5JE1
 
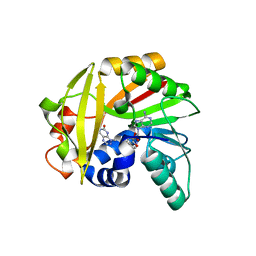 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
4JBS
 
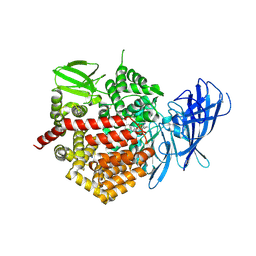 | | Crystal structure of the human Endoplasmic Reticulum Aminopeptidase 2 in complex with PHOSPHINIC PSEUDOTRIPEPTIDE inhibitor. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoplasmic reticulum aminopeptidase 2, ... | | Authors: | Saridakis, E, Birtley, J, Stratikos, E, Mavridis, I.M. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.789 Å) | | Cite: | Rationally designed inhibitor targeting antigen-trimming aminopeptidases enhances antigen presentation and cytotoxic T-cell responses.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7N2W
 
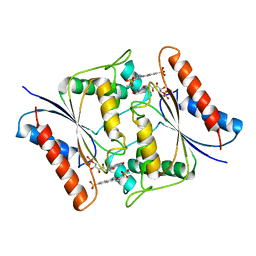 | | The crystal structure of an FMN-dependent NADH-azoreductase, AzoA in complex with Red 40 | | Descriptor: | 6-hydroxy-5-[(E)-(2-methoxy-5-methyl-4-sulfophenyl)diazenyl]naphthalene-2-sulfonic acid, FLAVIN MONONUCLEOTIDE, FMN dependent NADH:quinone oxidoreductase | | Authors: | Arcinas, A.J, Fedorov, E, Kelly, L, Almo, S.C, Ghosh, A. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Uncovering a novel mechanism of enzyme activation in multimeric azoreductases
To Be Published
|
|
6CUZ
 
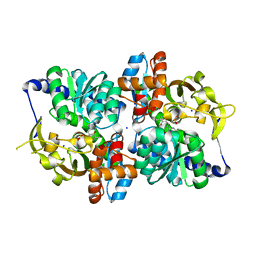 | | Engineered TrpB from Pyrococcus furiosus, PfTrpB7E6 with (2S,3R)-ethylserine bound as the amino-acrylate | | Descriptor: | (2E)-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pent-2-enoic acid, PHOSPHATE ION, SODIUM ION, ... | | Authors: | Scheele, R.A, Buller, A.R, Boville, C.E, Arnold, F.H. | | Deposit date: | 2018-03-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Engineered Biosynthesis of beta-Alkyl Tryptophan Analogues.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
3TAE
 
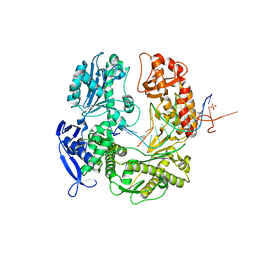 | | 5-hydroxycytosine paired with dAMP in RB69 gp43 | | Descriptor: | DNA (5'-D(*CP*CP*(5OC)P*GP*GP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*CP*CP*A)-3'), DNA polymerase, ... | | Authors: | Zahn, K.E. | | Deposit date: | 2011-08-04 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The miscoding potential of 5-hydroxycytosine arises due to template instability in the replicative polymerase active site.
Biochemistry, 50, 2011
|
|
7SQY
 
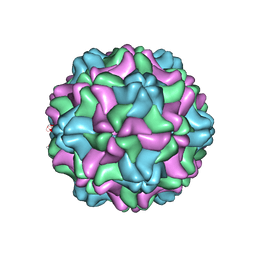 | | CSDaV GFP mutant | | Descriptor: | Citrus Sudden Death-associated Virus Capsid Protein,Green fluorescent protein,Citrus Sudden Death-associated Virus Capsid Protein | | Authors: | Guo, F, Matsumura, E.E, Falk, B.W. | | Deposit date: | 2021-11-07 | | Release date: | 2022-05-25 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Citrus sudden death-associated virus as a new expression vector for rapid in planta production of heterologous proteins, chimeric virions, and virus-like particles.
Biotechnol Rep., 35, 2022
|
|
6VXW
 
 | |
5AEC
 
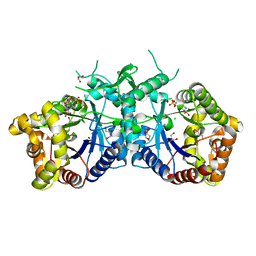 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2015-08-28 | | Release date: | 2015-09-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7DUL
 
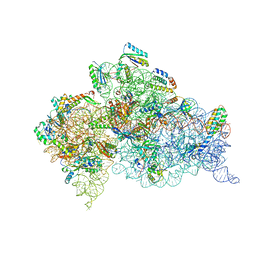 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N3''MS bound | | Descriptor: | (1S,2S,3R,4S,6R)-4,6-diamino-3-{[(2S,3R)-3-amino-6-(aminomethyl)-3,4-dihydro-2H-pyran-2-yl]oxy}-2-hydroxycyclohexyl 3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranoside, 30S Ribosomal RNA rRNA, 30S ribosomal protein S10, ... | | Authors: | DeMirci, H, Destan, E. | | Deposit date: | 2021-01-09 | | Release date: | 2022-01-26 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with mRNA and cognate transfer RNA anticodon stem-loop and sisomicin derivative N3''MS bound
To Be Published
|
|
4NXN
 
 | | Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8), bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2013-12-09 | | Release date: | 2014-05-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.544 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
4RM9
 
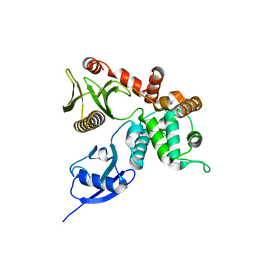 | | Crystal structure of human ezrin in space group C2221 | | Descriptor: | Ezrin | | Authors: | Phang, J.M, Harrop, S.J, Davies, R, Duff, A.P, Wilk, K.E, Curmi, P.M.G. | | Deposit date: | 2014-10-21 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization suggests models for monomeric and dimeric forms of full-length ezrin.
Biochem. J., 473, 2016
|
|
7TTL
 
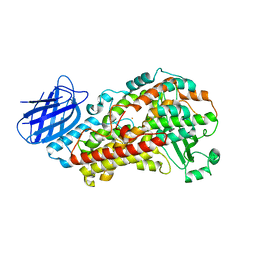 | | Stable-5-LOX elongated Ha2 (4 copies ASU) | | Descriptor: | Arachidonate 5-lipoxygenase, FE (II) ION | | Authors: | Gilbert, N.C, Newcomer, M.E. | | Deposit date: | 2022-02-01 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Helical remodeling augments 5-lipoxygenase activity in the synthesis of proinflammatory mediators.
J.Biol.Chem., 298, 2022
|
|
3MW6
 
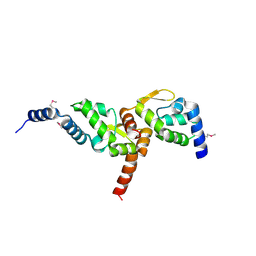 | | Crystal structure of NMB1681 from Neisseria meningitidis MC58, a FinO-like RNA chaperone | | Descriptor: | GLYCEROL, uncharacterized protein NMB1681 | | Authors: | Tan, K, Zhou, M, Duggan, E, Abdullah, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-05 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | N. meningitidis 1681 is a member of the FinO family of RNA chaperones.
Rna Biol., 7, 2010
|
|
3MWG
 
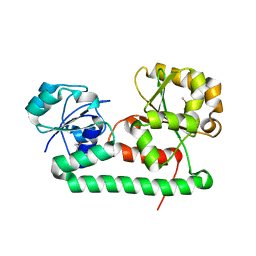 | | Crystal structure of Staphylococcus aureus SirA | | Descriptor: | Iron-regulated ABC transporter siderophore-binding protein SirA | | Authors: | Grigg, J.C, Murphy, M.E.P. | | Deposit date: | 2010-05-05 | | Release date: | 2010-09-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Staphylococcus aureus SirA specificity for staphyloferrin B is driven by localized conformational change
To be Published
|
|
5JDY
 
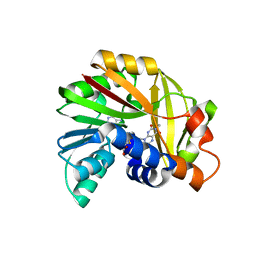 | | Crystal structure of Burkholderia glumae ToxA Y7F mutant with bound S-adenosylhomocysteine (SAH) and toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Methyl transferase, ... | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
1PTO
 
 | |
4S3D
 
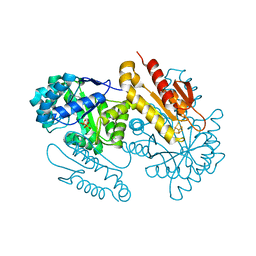 | | IspG in complex with PPi | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Quitterer, F, Frank, A, Wang, K, Rao, G, O'Dowd, B, Li, J, Guerra, F, Abdel-Azeim, S, Bacher, A, Eppinger, J, Oldfield, E, Groll, M. | | Deposit date: | 2015-01-26 | | Release date: | 2015-04-29 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE).
J.Mol.Biol., 427, 2015
|
|
5J4V
 
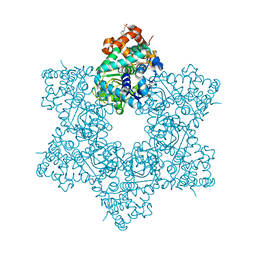 | | The crystal structure of Inhibitor Bound to JCV Helicase | | Descriptor: | 2-(2-phenoxypyridin-3-yl)[1,3]thiazolo[5,4-c]pyridine, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Large T antigen, ... | | Authors: | Ter Haar, E. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Fragment-Based Discovery of Dual JC Virus and BK Virus Helicase Inhibitors.
J.Med.Chem., 59, 2016
|
|
4RWB
 
 | | Racemic influenza M2-TM crystallized from monoolein lipidic cubic phase | | Descriptor: | Matrix protein 2, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Mortenson, D.E, Steinkruger, J.D, Kreitler, D.F, Gellman, S.H, Forest, K.T. | | Deposit date: | 2014-12-02 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of a heterochiral coiled coil.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2VZI
 
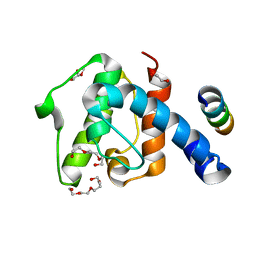 | | Crystal structure of the C-terminal calponin homology domain of alpha- parvin in complex with paxillin LD4 motif | | Descriptor: | 1,2-ETHANEDIOL, Alpha-parvin, Paxillin,Paxillin, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-08-01 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of the interactions between paxillin LD motifs and alpha-parvin.
Structure, 16, 2008
|
|
7TIN
 
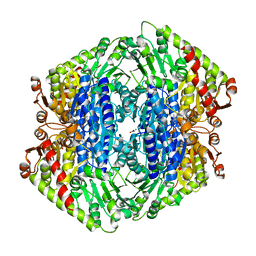 | | The Structure of S. aureus MenD | | Descriptor: | 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Johnston, J.M, Stanborough, T, Ho, N.A.T, Akazong, E.W, Jiao, W. | | Deposit date: | 2022-01-14 | | Release date: | 2022-09-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Allosteric inhibition of Staphylococcus aureus MenD by 1,4-dihydroxy naphthoic acid: a feedback inhibition mechanism of the menaquinone biosynthesis pathway.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 378, 2023
|
|
5KPX
 
 | | Structure of RelA bound to ribosome in presence of A/R tRNA (Structure IV) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Bah, E, Madireddy, R, Zhang, Y, Brilot, A.F, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-07-05 | | Release date: | 2016-09-28 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Ribosome•RelA structures reveal the mechanism of stringent response activation.
Elife, 5, 2016
|
|
7ICD
 
 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
