6WL3
 
 | | preTCRbeta-pMHC complex crystal structure | | Descriptor: | ARG-GLY-TYR-LEU-TYR-GLN-GLY-LEU, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
6GMP
 
 | | CRYSTAL STRUCTURE OF THE PPIASE DOMAIN OF TBPAR42 | | Descriptor: | PARVULIN 42 | | Authors: | Hoenig, D, Rute, A, Hofmann, E, Bayer, P, Gasper, R. | | Deposit date: | 2018-05-28 | | Release date: | 2019-03-20 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Analysis of the 42 kDa Parvulin ofTrypanosoma brucei.
Biomolecules, 9, 2019
|
|
6X4T
 
 | | Crystal structure of ICOS-L in complex with Prezalumab and VNAR domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Rujas, E, Sicard, T, Julien, J.P. | | Deposit date: | 2020-05-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural characterization of the ICOS/ICOS-L immune complex reveals high molecular mimicry by therapeutic antibodies.
Nat Commun, 11, 2020
|
|
6RJ8
 
 | | Structure of the alpha-beta hydrolase CorS from Tabernathe iboga | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Farrow, S.C, Caputi, L, Kamileen, M.O, Bussey, K, Stevenson, C.E.M, Mundy, J, Lawson, D.M, O'Connor, S.E. | | Deposit date: | 2019-04-26 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural basis of cycloaddition in biosynthesis of iboga and aspidosperma alkaloids.
Nat.Chem.Biol., 16, 2020
|
|
4Y9X
 
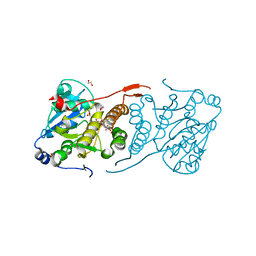 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and phosphoglyceric acid (PGA) - GpgS Mn2+ UDP-Glc PGA-3 | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, CHLORIDE ION, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6RV4
 
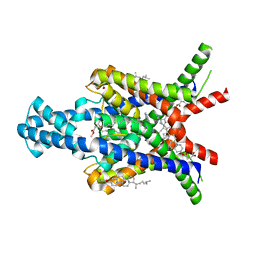 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation with a bound inhibitor BAY 2341237 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, POTASSIUM ION, ... | | Authors: | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-05-30 | | Release date: | 2019-08-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
6GS2
 
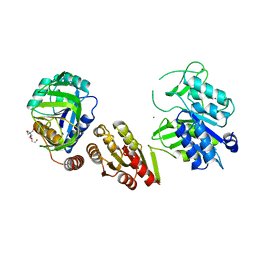 | | Crystal Structure of the GatD/MurT Enzyme Complex from Staphylococcus aureus | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, SA1707 protein, ... | | Authors: | Muckenfuss, L.M, Noeldeke, E.R, Niemann, V, Zocher, G, Stehle, T. | | Deposit date: | 2018-06-13 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural basis of cell wall peptidoglycan amidation by the GatD/MurT complex of Staphylococcus aureus.
Sci Rep, 8, 2018
|
|
5WCB
 
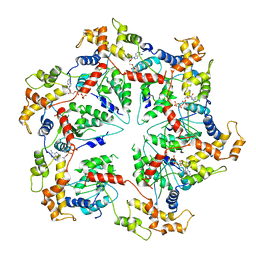 | | Katanin hexamer in the ring conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Meiotic spindle formation protein mei-1 | | Authors: | Zehr, E.A, Szyk, A, Piszczek, G, Szczesna, E, Zuo, X, Roll-Mecak, A. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Katanin spiral and ring structures shed light on power stroke for microtubule severing.
Nat. Struct. Mol. Biol., 24, 2017
|
|
6H65
 
 | | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Boyko, K.M, Timofeev, V.I, Bezsudnova, E.Y, Nikolaeva, A.Y, Rakitina, T.V, Popov, V.O. | | Deposit date: | 2018-07-26 | | Release date: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the branched-chain-amino-acid aminotransferase from Haliangium ochraceum
To Be Published
|
|
6Z7Z
 
 | |
4YST
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 24.9 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
6WKX
 
 | | Cryo-EM of Form 1 related peptide filament, 15-10-3 | | Descriptor: | peptide 15-10-3 | | Authors: | Wang, F, Gnewou, O.M, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
6WMF
 
 | | Human Sun2-KASH5 complex | | Descriptor: | POTASSIUM ION, Protein KASH5, SUN domain-containing protein 2 | | Authors: | Cruz, V.E, Schwartz, T.U. | | Deposit date: | 2020-04-21 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes.
J.Mol.Biol., 432, 2020
|
|
6TA1
 
 | | Fatty acid synthase of S. cerevisiae | | Descriptor: | FLAVIN MONONUCLEOTIDE, Fatty acid synthase subunit alpha, Fatty acid synthase subunit beta, ... | | Authors: | Vonck, J, D'Imprima, E, Joppe, M, Grininger, M. | | Deposit date: | 2019-10-29 | | Release date: | 2019-11-06 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The resolution revolution in cryoEM requires high-quality sample preparation: a rapid pipeline to a high-resolution map of yeast fatty acid synthase.
Iucrj, 7, 2020
|
|
8OW6
 
 | |
1Y4D
 
 | | Crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59R/E60S mutant | | Descriptor: | CALCIUM ION, SODIUM ION, chymotrypsin inhibitor 2, ... | | Authors: | Radisky, E.S, Lu, C.J, Kwan, G, Koshland Jr, D.E. | | Deposit date: | 2004-11-30 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Role of the intramolecular hydrogen bond network in the inhibitory power of chymotrypsin inhibitor 2
Biochemistry, 44, 2005
|
|
4Z2B
 
 | | The structure of human PDE12 residues 161-609 in complex with GSK3036342A | | Descriptor: | 1,2-ETHANEDIOL, 2',5'-phosphodiesterase 12, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nolte, R.T, Wisely, B, Wang, L, Wood, E.R. | | Deposit date: | 2015-03-29 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Role of Phosphodiesterase 12 (PDE12) as a Negative Regulator of the Innate Immune Response and the Discovery of Antiviral Inhibitors.
J.Biol.Chem., 290, 2015
|
|
4Z3U
 
 | | PRV nuclear egress complex | | Descriptor: | CHLORIDE ION, SODIUM ION, UL31, ... | | Authors: | Bigalke, J.M, Heldwein, E.E. | | Deposit date: | 2015-03-31 | | Release date: | 2015-11-11 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.706 Å) | | Cite: | Structural basis of membrane budding by the nuclear egress complex of herpesviruses.
Embo J., 34, 2015
|
|
6TMP
 
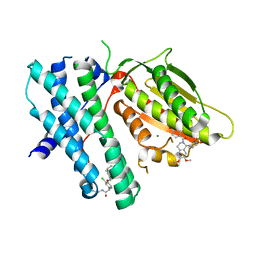 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [2,4-bis(oxidanyl)phenyl]-[(1~{R})-6,7-dimethoxy-1-pyridin-3-yl-3,4-dihydro-1~{H}-isoquinolin-2-yl]methanone, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.076 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
3GJE
 
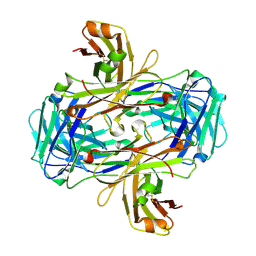 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Fab Heavy Chain, Fab Light Chain | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E, Kleber, S, Stenner-Liewen, F, Bauer, S, McMichael, A, Knuth, A, Abken, H, Hombach, A.A, Cerundolo, V, Jones, E.Y, Renner, C. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
8VU6
 
 | |
6GCU
 
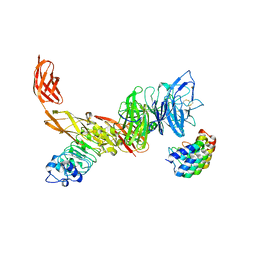 | | MET receptor in complex with InlB internalin domain and DARPin A3A | | Descriptor: | DARPin A3A, Hepatocyte growth factor receptor, Internalin B | | Authors: | Meyer, T, Andres, F, Iamele, L, Gherardi, E, Pluckthun, A, Niemann, H.H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (6.001 Å) | | Cite: | Inhibition of the MET Kinase Activity and Cell Growth in MET-Addicted Cancer Cells by Bi-Paratopic Linking.
J.Mol.Biol., 431, 2019
|
|
6G9B
 
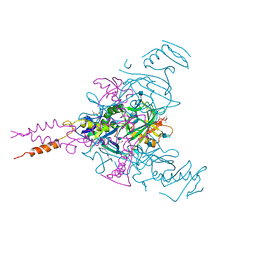 | | Crystal structure of Ebolavirus glycoprotein in complex with imipramine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5H-DIBENZO[B,F]AZEPIN-5-YL)-N,N-DIMETHYLPROPAN-1-AMINE, ... | | Authors: | Zhao, Y, Ren, J, Fry, E.E, Xiao, J, Townsend, A.R, Stuart, D.I. | | Deposit date: | 2018-04-10 | | Release date: | 2018-05-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structures of Ebola Virus Glycoprotein Complexes with Tricyclic Antidepressant and Antipsychotic Drugs.
J. Med. Chem., 61, 2018
|
|
8VU8
 
 | |
6WL1
 
 | | Cryo-EM of Form 1 related peptide filament, 36-31-3 | | Descriptor: | peptide 36-31-3 | | Authors: | Wang, F, Gnewou, O.M, Modlin, C, Egelman, E.H, Conticello, V.P. | | Deposit date: | 2020-04-17 | | Release date: | 2020-12-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural analysis of cross alpha-helical nanotubes provides insight into the designability of filamentous peptide nanomaterials.
Nat Commun, 12, 2021
|
|
