8UTB
 
 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine and NS-1738 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Noviello, C.M, Hibbs, R.E. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
5B6Y
 
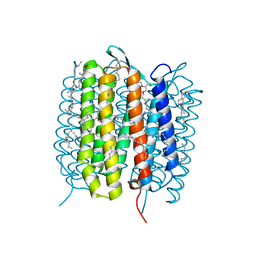 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 36.2 us after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
4AN5
 
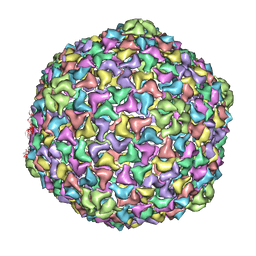 | | Capsid structure and its Stability at the Late Stages of Bacteriophage SPP1 Assembly | | Descriptor: | COAT PROTEIN | | Authors: | White, H.E, Sherman, M.B, Brasiles, S, Jacquet, E, Seavers, P, Tavares, P, Orlova, E.V. | | Deposit date: | 2012-03-15 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Capsid Structure and its Stability at the Late Stages of Bacteriophage Spp1 Assembly.
J.Virol., 86, 2012
|
|
8UT1
 
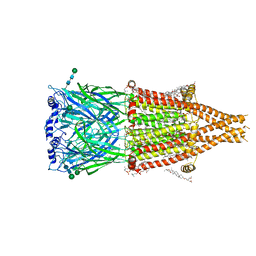 | | Alpha7-nicotinic acetylcholine receptor bound to epibatidine | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Burke, S.M, Hibbs, R.E, Noviello, C.M. | | Deposit date: | 2023-10-30 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural mechanisms of alpha 7 nicotinic receptor allosteric modulation and activation.
Cell, 187, 2024
|
|
2AV0
 
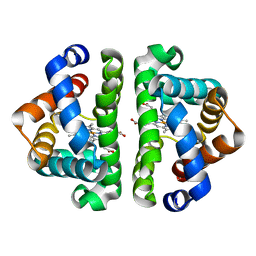 | | F97L with CO bound | | Descriptor: | CARBON MONOXIDE, Globin I, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Knapp, J.E, Bonham, M.A, Gibson, Q.H, Nichols, J.C, Royer Jr, W.E. | | Deposit date: | 2005-08-29 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Residue F4 plays a key role in modulating oxygen affinity and cooperativity in Scapharca dimeric hemoglobin
Biochemistry, 44, 2005
|
|
8P07
 
 | | Crystal structure of human Casein Kinase II subunit alpha (CK2a1) in complex with 5-((3-(4H-1,2,4-triazol-4-yl)phenyl)amino)-7-(cyclopropylamino)pyrazolo[1,5-a]pyrimidine-3-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 7-(cyclopropylamino)-5-[[3-(1,2,4-triazol-4-yl)phenyl]amino]pyrazolo[1,5-a]pyrimidine-3-carbonitrile, Casein kinase II subunit alpha, ... | | Authors: | Kraemer, A, Ong, H.W, Yang, X, Bown, J.W, Chang, E, Willson, T, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-05-09 | | Release date: | 2023-05-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | More than an Amide Bioisostere: Discovery of 1,2,4-Triazole-containing Pyrazolo[1,5- a ]pyrimidine Host CSNK2 Inhibitors for Combatting beta-Coronavirus Replication.
J.Med.Chem., 67, 2024
|
|
4U8A
 
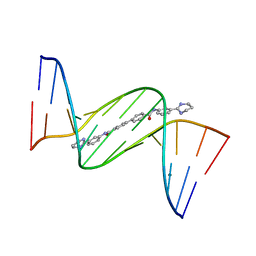 | | Crystal structure of D(CGCGAATTCGCG)2 complexed with BPH-1503 | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION, N,N'-bis[3-(1,4,5,6-tetrahydropyrimidin-2-yl)phenyl]biphenyl-4,4'-dicarboxamide | | Authors: | Zhu, W, Oldfield, E. | | Deposit date: | 2014-08-01 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Antibacterial drug leads: DNA and enzyme multitargeting.
J.Med.Chem., 58, 2015
|
|
8OM9
 
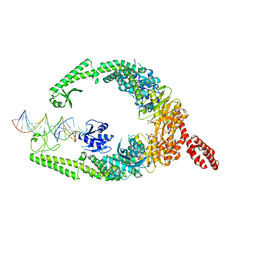 | | MutSbeta bound to (CAG)2 DNA (open form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (open form)
To Be Published
|
|
4TW1
 
 | | Crystal structure of the octameric pore complex of the Staphylococcus aureus Bi-component Toxin LukGH | | Descriptor: | Possible leukocidin subunit | | Authors: | Logan, D.T, Hakansson, M, Saline, M, Kimbung, R, Badarau, A, Rouha, H, Nagy, E. | | Deposit date: | 2014-06-29 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Function Analysis of Heterodimer Formation, Oligomerization, and Receptor Binding of the Staphylococcus aureus Bi-component Toxin LukGH.
J.Biol.Chem., 290, 2015
|
|
8ONO
 
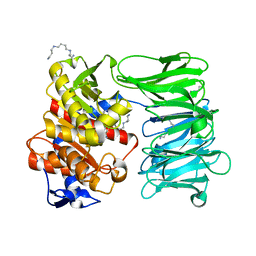 | | Modified oligopeptidase B from S. proteamaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution | | Descriptor: | Oligopeptidase B, SPERMINE | | Authors: | Petrenko, D.E, Boyko, K.M, Nikolaeva, A.Y, Vlaskina, A.V, Mikhailova, A.G, Timofeev, V.I, Rakitina, T.V. | | Deposit date: | 2023-04-03 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Modified oligopeptidase B from S. proteomaculans in intermediate conformation with 5 spermine molecule at 1.65 A resolution
To Be Published
|
|
8Q9Q
 
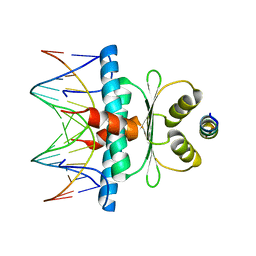 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC7 deacetylase binding motif | | Descriptor: | HDAC7 (histone deacetylase 7) binding motif peptide: GLY-VAL-VAL-LYS-GLN-LYS-LEU-ALA-GLU-VAL-ILE-LEU-LYS-LYS-GLN, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
4FX6
 
 | | Crystal structure of the mutant V182A.R203A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2012-07-02 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.531 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
8OLX
 
 | | MutSbeta bound to (CAG)2 DNA (canonical form) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (25-MER), DNA mismatch repair protein Msh2, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-30 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | MutSbeta bound to (CAG)2 DNA (canonical form)
To Be Published
|
|
1RDZ
 
 | | T-STATE STRUCTURE OF THE ARG 243 TO ALA MUTANT OF PIG KIDNEY FRUCTOSE 1,6-BISPHOSPHATASE EXPRESSED IN E. COLI | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE 1,6-BISPHOSPHATASE | | Authors: | Stec, B, Abraham, R, Giroux, E, Kantrowitz, E.R. | | Deposit date: | 1996-05-17 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structures of the active site mutant (Arg-243-->Ala) in the T and R allosteric states of pig kidney fructose-1,6-bisphosphatase expressed in Escherichia coli.
Protein Sci., 5, 1996
|
|
7V5L
 
 | | Crystal structure of human bleomycin hydrolase | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
5J6C
 
 | | FMN-dependent Nitroreductase (CDR20291_0767) from Clostridium difficile R20291 | | Descriptor: | FLAVIN MONONUCLEOTIDE, IMIDAZOLE, Putative reductase | | Authors: | Powell, S.M, Wang, B, Hessami, N, Najar, F.Z, Thomas, L.M, West, A.H, Karr, E.A, Richter-Addo, G.B. | | Deposit date: | 2016-04-04 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Crystal structures of two nitroreductases from hypervirulent Clostridium difficile and functionally related interactions with the antibiotic metronidazole.
Nitric Oxide, 60, 2016
|
|
4ZGQ
 
 | | Structure of Cdc123 bound to eIF2-gammaDIII domain | | Descriptor: | Cell division cycle protein 123, Eukaryotic translation initiation factor 2 subunit gamma | | Authors: | Panvert, M, Dubiez, E, Arnold, L, Perez, J, Seufert, W, Mechulam, Y, Schmitt, E. | | Deposit date: | 2015-04-23 | | Release date: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Cdc123, a Cell Cycle Regulator Needed for eIF2 Assembly, Is an ATP-Grasp Protein with Unique Features.
Structure, 23, 2015
|
|
6CCV
 
 | | Crystal structure of a Mycobacterium smegmatis RNA polymerase transcription initiation complex with inhibitor Rifampicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Rifamycin congeners kanglemycins are active against rifampicin-resistant bacteria via a distinct mechanism.
Nat Commun, 9, 2018
|
|
8OMO
 
 | | DNA-unbound MutSbeta-ATP complex (bent clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (bent clamp form)
To Be Published
|
|
5DW7
 
 | | Crystal structure of the unliganded geosmin synthase N-terminal domain from Streptomyces coelicolor | | Descriptor: | Germacradienol/geosmin synthase | | Authors: | Lombardi, P.M, Harris, G.G, Pemberton, T.A, Matsui, T, Weiss, T.M, Cole, K.E, Koksal, M, Murphy, F.V, Vedula, L.S, Chou, W.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2015-09-22 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural Studies of Geosmin Synthase, a Bifunctional Sesquiterpene Synthase with alpha alpha Domain Architecture That Catalyzes a Unique Cyclization-Fragmentation Reaction Sequence.
Biochemistry, 54, 2015
|
|
8OM5
 
 | | DNA-free open form of MutSbeta | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | DNA-free open form of MutSbeta
To Be Published
|
|
8OMA
 
 | | MutSbeta bound to 61bp homoduplex DNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | MutSbeta bound to 61bp homoduplex DNA
To Be Published
|
|
8OMQ
 
 | | DNA-unbound MutSbeta-ATP complex (straight clamp form) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA mismatch repair protein Msh2, DNA mismatch repair protein Msh3, ... | | Authors: | Lee, J.-H, Thomsen, M, Daub, H, Steinbacher, S, Sztyler, A, Thieulin-Pardo, G, Neudegger, T, Plotnikov, N, Iyer, R.R, Wilkinson, H, Monteagudo, E, Felsenfeld, D.P, Haque, T, Finley, M, Dominguez, C, Vogt, T.F, Prasad, B.C. | | Deposit date: | 2023-03-31 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | DNA-unbound MutSbeta-ATP complex (straight clamp form)
To Be Published
|
|
1D4M
 
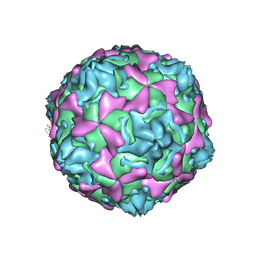 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
5TCY
 
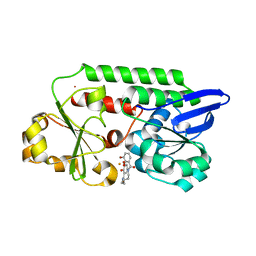 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
