6EWK
 
 | | T. californica AChE in complex with a 3-hydroxy-2-pyridine aldoxime. | | Descriptor: | 2-[(~{E})-hydroxyiminomethyl]-6-(5-morpholin-4-ylpentyl)pyridin-3-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | de la Mora, E, Weik, M, Braiki, A, Mougeot, R, Jean, L, Renard, P.I. | | Deposit date: | 2017-11-04 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Potent 3-Hydroxy-2-Pyridine Aldoxime Reactivators of Organophosphate-Inhibited Cholinesterases with Predicted Blood-Brain Barrier Penetration.
Chemistry, 24, 2018
|
|
8JVG
 
 | | Crystal structure of dephospho-coenzyme A kinase | | Descriptor: | GTP-dependent dephospho-CoA kinase | | Authors: | Kita, A, Ishida, Y, Shimosaka, T, Michimori, Y, Makarova, K, Koonin, E, Atomi, H, Miki, K. | | Deposit date: | 2023-06-28 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of GTP-dependent dephospho-coenzyme A kinase from the hyperthermophilic archaeon, Thermococcus kodakarensis.
Proteins, 92, 2024
|
|
6SQM
 
 | | Crystal structure of CREBBP bromodomain complexed with LA36 | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, ~{N}-[3-acetamido-5-[(3-methylcinnolin-5-yl)carbamoyl]phenyl]furan-2-carboxamide | | Authors: | Bedi, R.K, Laul, E, Nevado, C, Caflisch, A. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of CREBBP bromodomain complexed with LA36
To Be Published
|
|
6H1F
 
 | | Structure of the nanobody-stabilized gelsolin D187N variant (second domain) | | Descriptor: | Gelsolin, THIOCYANATE ION, gelsolin nanobody, ... | | Authors: | Hassan, A, Milani, M, Mastrangelo, E, de Rosa, M. | | Deposit date: | 2018-07-11 | | Release date: | 2019-01-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nanobody interaction unveils structure, dynamics and proteotoxicity of the Finnish-type amyloidogenic gelsolin variant.
Biochim Biophys Acta Mol Basis Dis, 1865, 2019
|
|
6F2Q
 
 | | Neutron crystal structure of perdeuterated galectin-3C in the ligand-free form | | Descriptor: | Galectin-3 | | Authors: | Manzoni, F, Blakeley, M.P, Oksanen, E, Logan, D.T. | | Deposit date: | 2017-11-27 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.03 Å), X-RAY DIFFRACTION | | Cite: | Elucidation of Hydrogen Bonding Patterns in Ligand-Free, Lactose- and Glycerol-Bound Galectin-3C by Neutron Crystallography to Guide Drug Design.
J. Med. Chem., 61, 2018
|
|
3JZU
 
 | | Crystal structure of Dipeptide Epimerase from Enterococcus faecalis V583 complexed with Mg and dipeptide L-Leu-L-Tyr | | Descriptor: | Dipeptide Epimerase, LEUCINE, MAGNESIUM ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Imker, H.J, Sakai, A, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-09-24 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7N5K
 
 | | PCNA from Thermococcus gammatolerans: crystal II, collection 1, 1.98 A, 3.84 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5J
 
 | | PCNA from Thermococcus gammatolerans: crystal I, collection 5, 2.82 A, 89.1 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
6GGO
 
 | | Crystal structure of Salmonella zinc metalloprotease effector GtgA | | Descriptor: | Bacteriophage virulence determinant, CHLORIDE ION, ZINC ION | | Authors: | Jennings, E, Esposito, D, Rittinger, K, Thurston, T. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-function analyses of the bacterial zinc metalloprotease effector protein GtgA uncover key residues required for deactivating NF-kappa B.
J. Biol. Chem., 293, 2018
|
|
7N5M
 
 | | PCNA from Thermococcus gammatolerans: crystal III, collection 1, 2.00 A, 1.91 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5I
 
 | | PCNA from Thermococcus gammatolerans: crystal I, collection 1, 1.95 A, 5.22 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
1VKC
 
 | | Putative acetyl transferase from Pyrococcus furiosus | | Descriptor: | IODIDE ION, putative acetyl transferase | | Authors: | Habel, J.E, Liu, Z.-J, Tempel, W, Rose, J.P, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Poole II, F.L, Shah, C, Sugar, F.J, Adams, M.W.W, Richardson, D.C, Richardson, J.S, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Putative acetyl transferase from Pyrococcus furiosus
To be published
|
|
7N5N
 
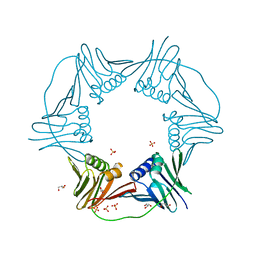 | | PCNA from Thermococcus gammatolerans: crystal III, collection 15, 2.20 A, 28.7 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
1AT1
 
 | |
2GU2
 
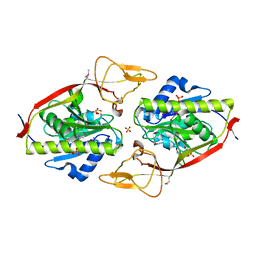 | | Crystal Structure of an Aspartoacylase from Rattus norvegicus | | Descriptor: | Aspa protein, SULFATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-04-28 | | Release date: | 2006-06-20 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7N5L
 
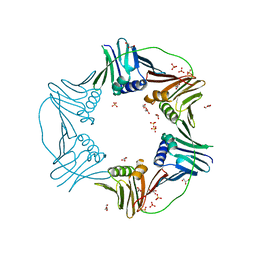 | | PCNA from Thermococcus gammatolerans: crystal II, collection 20, 3.07 A, 77.0 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
5JNV
 
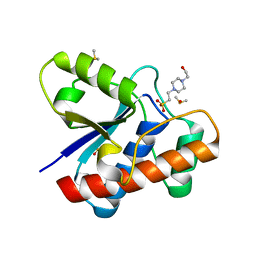 | | Crystal structure of bovine low molecular weight protein tyrosine phosphatase (LMPTP) mutant (W49Y N50E) complexed with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, Low molecular weight phosphotyrosine protein phosphatase | | Authors: | Stanford, S.M, Aleshin, A.E, Liddington, R.C, Bankston, L, Cadwell, G, Bottini, N. | | Deposit date: | 2016-04-30 | | Release date: | 2017-03-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diabetes reversal by inhibition of the low-molecular-weight tyrosine phosphatase.
Nat. Chem. Biol., 13, 2017
|
|
7TOO
 
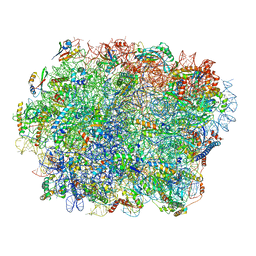 | | Yeast 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein GR20 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
3K77
 
 | | X-ray crystal structure of XRCC1 | | Descriptor: | DNA repair protein XRCC1 | | Authors: | Cuneo, M.J, London, R.E. | | Deposit date: | 2009-10-12 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Oxidation state of the XRCC1 N-terminal domain regulates DNA polymerase beta binding affinity.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6VH6
 
 | |
6SWD
 
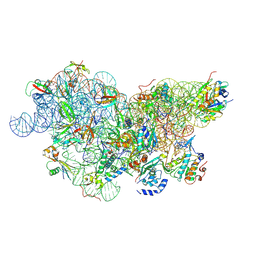 | | IC2 body model of cryo-EM structure of a full archaeal ribosomal translation initiation complex devoid of aIF1 in P. abyssi | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Coureux, P.-D, Mechulam, Y, Schmitt, E. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM study of an archaeal 30S initiation complex gives insights into evolution of translation initiation.
Commun Biol, 3, 2020
|
|
5NG1
 
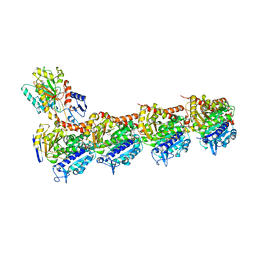 | | TUBULIN-MTC-zampanolide complex | | Descriptor: | (2Z,4E)-N-[(S)-[(1S,2E,5S,8E,10Z,17S)-3,11-dimethyl-19-methylidene-7,13-dioxo-6,21-dioxabicyclo[15.3.1]henicosa-2,8,10-trien-5-yl](hydroxy)methyl]hexa-2,4-dienamide, (2~{Z},4~{E})-~{N}-[(~{S})-oxidanyl-[(1~{S},2~{E},5~{S},11~{R},17~{S},19~{R})-3,11,19-trimethyl-7,13-bis(oxidanylidene)-6,21-dioxabicyclo[15.3.1]henicos-2-en-5-yl]methyl]hexa-2,4-dienamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Field, J.J, Pera, B, Estevez Gallego, J, Calvo, E, Rodriguez-Salarichs, J, Saez-Calvo, G, Zuwerra, D, Jordi, M, Prota, A.E, Menchon, G, Miller, J.H, Altmann, K.-H, Diaz, J.F. | | Deposit date: | 2017-03-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Zampanolide Binding to Tubulin Indicates Cross-Talk of Taxane Site with Colchicine and Nucleotide Sites.
J. Nat. Prod., 81, 2018
|
|
6GOZ
 
 | | Structure of mEos4b in the green long-lived dark state | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | De Zitter, E, Adam, V, Byrdin, M, Van Meervelt, L, Dedecker, P, Bourgeois, D. | | Deposit date: | 2018-06-04 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.406 Å) | | Cite: | Mechanistic Investigations of Green mEos4b Reveal a Dynamic Long-Lived Dark State.
J.Am.Chem.Soc., 2020
|
|
6SPD
 
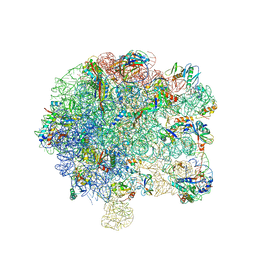 | | Pseudomonas aeruginosa 50s ribosome from a clinical isolate | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L11, 50S ribosomal protein L13, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-16 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7TOP
 
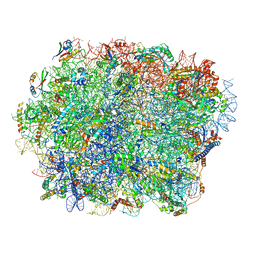 | | Yeast 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein PR20 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
