2YAD
 
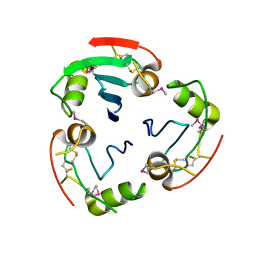 | | BRICHOS domain of Surfactant protein C precursor protein | | Descriptor: | SURFACTANT PROTEIN C BRICHOS DOMAIN | | Authors: | Askarieh, G, Siponen, M.I, Willander, H, Landreh, M, Westermark, P, Nordling, K, Keranen, H, Hermansson, E, Hamvas, A, Nogee, L.M, Bergman, T, Saenz, A, Casals, C, Aqvist, J, Jornvall, H, Presto, J, Johansson, J, Arrowsmith, C.H, Bountra, C, Collins, R, Edwards, A.M, Ekblad, T, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Karlberg, T, Kol, S, Kotenyova, T, Kouznetsova, E, Moche, M, Nyman, T, Nordlund, P, Persson, C, Schuler, H, Thorsell, A.G, Tresaugues, L, van den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Berglund, H, Knight, S.D. | | Deposit date: | 2011-02-18 | | Release date: | 2012-02-15 | | Last modified: | 2014-08-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution Structure of a Bricos Domain and its Implications for Anti-Amyloid Chaperone Activity on Lung Surgactant Protein C.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8SSZ
 
 | |
1HY7
 
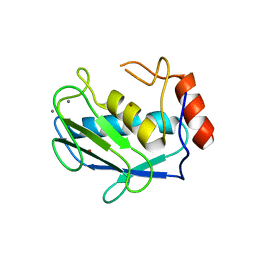 | | A CARBOXYLIC ACID BASED INHIBITOR IN COMPLEX WITH MMP3 | | Descriptor: | CALCIUM ION, R-2-{[4'-METHOXY-(1,1'-BIPHENYL)-4-YL]-SULFONYL}-AMINO-6-METHOXY-HEX-4-YNOIC ACID, STROMELYSIN-1, ... | | Authors: | Natchus, M.G, Bookland, R.G, Laufersweiler, M.J, Pikul, S, Almstead, N.G, De, B, Janusz, M.J, Hsieh, L.C, Gu, F, Pokross, M.E, Patel, V.S, Garver, S.M, Peng, S.X, Branch, T.M, King, S.L, Baker, T.R, Foltz, D.J, Mieling, G.E. | | Deposit date: | 2001-01-18 | | Release date: | 2002-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of new carboxylic acid-based MMP inhibitors derived from functionalized propargylglycines.
J.Med.Chem., 44, 2001
|
|
3KKK
 
 | |
5B6X
 
 | | A three dimensional movie of structural changes in bacteriorhodopsin: structure obtained 760 ns after photoexcitation | | Descriptor: | 2,3-DI-PHYTANYL-GLYCEROL, Bacteriorhodopsin, DECANE, ... | | Authors: | Royant, A, Nango, E, Nakane, T, Tanaka, T, Arima, T, Neutze, R, Iwata, S. | | Deposit date: | 2016-06-02 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A three-dimensional movie of structural changes in bacteriorhodopsin
Science, 354, 2016
|
|
1U1S
 
 | | Hfq protein from Pseudomonas aeruginosa. Low-salt crystals | | Descriptor: | Hfq protein | | Authors: | Nikulin, A.D, Stolboushkina, E.A, Perederina, I, Vassilieva, I.M, Blaesi, U, Moll, I, Kachalova, G, Vassylyev, D, Yokoyama, S, Garber, M, Nikonov, S.V, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Pseudomonas aeruginosa Hfq protein.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
5IQ5
 
 | | NMR solution structure of Mayaro virus macro domain | | Descriptor: | Macro domain | | Authors: | Melekis, E, Tsika, A.C, Bentrop, D, Papageorgiou, N, Coutard, B, Spyroulias, G.A. | | Deposit date: | 2016-03-10 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Deciphering the Nucleotide and RNA Binding Selectivity of the Mayaro Virus Macro Domain.
J.Mol.Biol., 431, 2019
|
|
4QA3
 
 | | Crystal structure of T311M HDAC8 in complex with Trichostatin A (TSA) | | Descriptor: | GLYCEROL, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Decroos, C, Bowman, C.B, Moser, J.-A.S, Christianson, K.E, Deardorff, M.A, Christianson, D.W. | | Deposit date: | 2014-05-02 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.876 Å) | | Cite: | Compromised Structure and Function of HDAC8 Mutants Identified in Cornelia de Lange Syndrome Spectrum Disorders.
Acs Chem.Biol., 9, 2014
|
|
2OPG
 
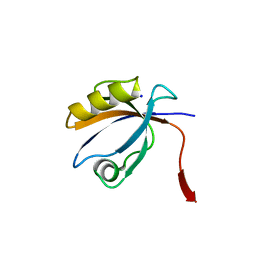 | | The crystal structure of the 10th PDZ domain of MPDZ | | Descriptor: | Multiple PDZ domain protein, SODIUM ION | | Authors: | Gileadi, C, Phillips, C, Elkins, J, Papagrigoriou, E, Ugochukwu, E, Gorrec, F, Savitsky, P, Umeano, C, Berridge, G, Gileadi, O, Salah, E, Edwards, A, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-01-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of the 10th PDZ domain of MPDZ
To be Published
|
|
7TLI
 
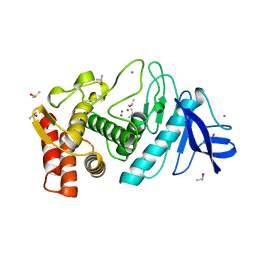 | | THERMOLYSIN (90% ISOPROPANOL SOAKED CRYSTALS) | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, ISOPROPYL ALCOHOL, ... | | Authors: | English, A.C, Done, S.H, Groom, C.R, Hubbard, R.E. | | Deposit date: | 1998-10-30 | | Release date: | 2000-03-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Locating interaction sites on proteins: the crystal structure of thermolysin soaked in 2% to 100% isopropanol.
Proteins, 37, 1999
|
|
7TLN
 
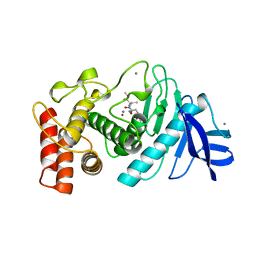 | | STRUCTURAL ANALYSIS OF THE INHIBITION OF THERMOLYSIN BY AN ACTIVE-SITE-DIRECTED IRREVERSIBLE INHIBITOR | | Descriptor: | 2-(ACETYL-HYDROXY-AMINO)-4-METHYL-PENTANOIC ACID METHYL ESTER, CALCIUM ION, THERMOLYSIN, ... | | Authors: | Matthews, B.W, Holmes, M.A, Tronrud, D.E. | | Deposit date: | 1983-01-27 | | Release date: | 1983-03-09 | | Last modified: | 2022-11-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of the inhibition of thermolysin by an active-site-directed irreversible inhibitor.
Biochemistry, 22, 1983
|
|
8PK0
 
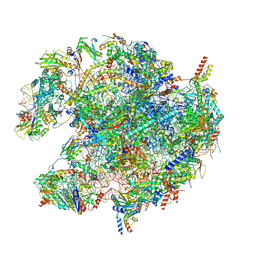 | | human mitoribosomal large subunit assembly intermediate 1 with GTPBP10-GTPBP7 | | Descriptor: | 16S rRNA + pre-H68-71 segment, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Kummer, E, Nguyen, T.G, Ritter, C. | | Deposit date: | 2023-06-23 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights into the role of GTPBP10 in the RNA maturation of the mitoribosome.
Nat Commun, 14, 2023
|
|
5IYA
 
 | | Human core-PIC in the closed state | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB10, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | He, Y, Yan, C, Fang, J, Inouye, C, Tjian, R, Ivanov, I, Nogales, E. | | Deposit date: | 2016-03-24 | | Release date: | 2016-05-18 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Near-atomic resolution visualization of human transcription promoter opening.
Nature, 533, 2016
|
|
5BPU
 
 | | Crystal structure of Norrin, a Wnt signalling activator, Crystal Form I | | Descriptor: | (GGL)EEE, (GGL)EEEEEE, Norrin | | Authors: | Chang, T.-H, Hsieh, F.-L, Harlos, K, Jones, E.Y. | | Deposit date: | 2015-05-28 | | Release date: | 2015-07-01 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and functional properties of Norrin mimic Wnt for signalling with Frizzled4, Lrp5/6, and proteoglycan.
Elife, 4, 2015
|
|
2OVP
 
 | | Structure of the Skp1-Fbw7 complex | | Descriptor: | F-box/WD repeat protein 7, S-phase kinase-associated protein 1A | | Authors: | Hao, B, Oehlmann, S, Sowa, M.E, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of a Fbw7-Skp1-Cyclin E Complex: Multisite-Phosphorylated Substrate Recognition by SCF Ubiquitin Ligases
Mol.Cell, 26, 2007
|
|
1LYB
 
 | | CRYSTAL STRUCTURES OF NATIVE AND INHIBITED FORMS OF HUMAN CATHEPSIN D: IMPLICATIONS FOR LYSOSOMAL TARGETING AND DRUG DESIGN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CATHEPSIN D, PEPSTATIN, ... | | Authors: | Baldwin, E.T, Bhat, T.N, Gulnik, S, Erickson, J.W. | | Deposit date: | 1993-04-22 | | Release date: | 1994-01-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design.
Proc.Natl.Acad.Sci.USA, 90, 1993
|
|
8I3Y
 
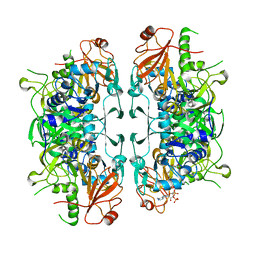 | | Crystal structure of ASCT from Trypanosoma brucei in complex with Succinyl-CoA. | | Descriptor: | CALCIUM ION, SUCCINIC ACID, SUCCINYL-COENZYME A, ... | | Authors: | Mochizuki, K, Inaoka, D.K, Fukuda, K, Kurasawa, H, Iyoda, K, Nakai, U, Harada, S, Balogun, E.O, Mazet, M, Millerioux, Y, Bringaud, F, Boshart, M, Hirayama, K, Kita, K, Shiba, T. | | Deposit date: | 2023-01-18 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of ASCT from Trypanosoma brucei in complex with Succinyl-CoA.
To Be Published
|
|
8TLC
 
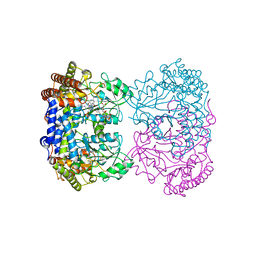 | | Human mitochondrial serine hydroxymethyltransferase (SHMT2) in complex with PLP, glycine and tri-glutamate AGF347 inhibitor | | Descriptor: | N-{4-[4-(2-amino-4-oxo-1,4-dihydro-5H-pyrrolo[3,2-d]pyrimidin-5-yl)butyl]-2-fluorobenzoyl}-D-gamma-glutamyl-L-gamma-glutamyl-D-glutamic acid, Serine hydroxymethyltransferase, mitochondrial | | Authors: | Katinas, J.M, Dann III, C.E. | | Deposit date: | 2023-07-26 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Characterization of 5-Substituted Pyrrolo[3,2- d ]pyrimidine Antifolate Inhibitors in Complex with Human Serine Hydroxymethyl Transferase 2.
Biochemistry, 2024
|
|
8TNR
 
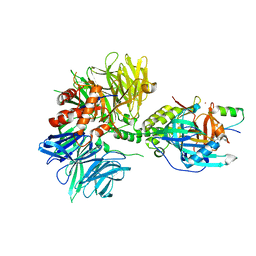 | | Cryo-EM structure of DDB1dB:CRBN:PT-179:SD40, conformation 2 | | Descriptor: | 2-[(3S)-2,6-dioxopiperidin-3-yl]-5-(morpholin-4-yl)-1H-isoindole-1,3(2H)-dione, DNA damage-binding protein 1, Maltose/maltodextrin-binding periplasmic protein,SD40, ... | | Authors: | Roy Burman, S.S, Hunkeler, M, Fischer, E.S. | | Deposit date: | 2023-08-02 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Continuous evolution of compact protein degradation tags regulated by selective molecular glues.
Science, 383, 2024
|
|
3O4L
 
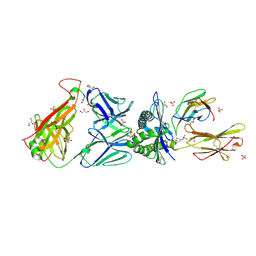 | | Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BSLF2/BMLF1 protein, Beta-2-microglobulin, ... | | Authors: | Miles, J.J, Bulek, A.M, Cole, D.K, Gostick, E, Schauenburg, J.A, Dolton, G, Venturi, V, Davenport, M.P, Tan, M.P, Burrows, S.R, Wooldridge, L, Price, D.A, Rizkallah, P.J, Sewell, A.K. | | Deposit date: | 2010-07-27 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Genetic and structural basis for selection of a ubiquitous T cell receptor deployed in Epstein-Barr virus infection.
Plos Pathog., 6, 2010
|
|
5J1A
 
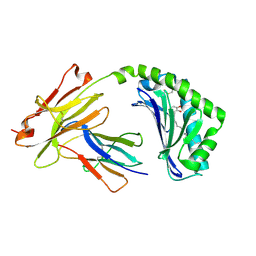 | | Antigen presenting molecule | | Descriptor: | 3-[(8Z,11Z)-pentadeca-8,11-dien-1-yl]benzene-1,2-diol, Beta-2-microglobulin, T-cell surface glycoprotein CD1a | | Authors: | Yongqing, T, Rossjohn, J, Le Nours, J, Marquez, E, Winau, F. | | Deposit date: | 2016-03-29 | | Release date: | 2016-08-03 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | CD1a on Langerhans cells controls inflammatory skin disease.
Nat. Immunol., 17, 2016
|
|
5AOG
 
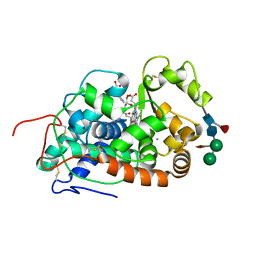 | | Structure of Sorghum peroxidase | | Descriptor: | 1H-INDOL-3-YLACETIC ACID, CALCIUM ION, CATIONIC PEROXIDASE SPC4, ... | | Authors: | Kwon, H, Nnamchi, C.I, Parkin, G, Efimov, I, Agirre, J, Basran, J, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2015-09-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural and Spectroscopic Characterisation of a Heme Peroxidase from Sorghum.
J.Biol.Inorg.Chem., 21, 2016
|
|
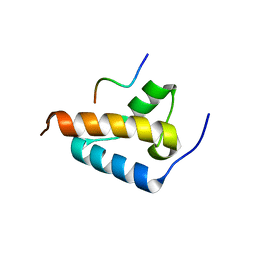
8TGC
 
 | |
5ARG
 
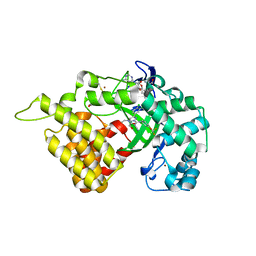 | | SMYD2 in complex with SGC probe BAY-598 | | Descriptor: | GLYCEROL, N-LYSINE METHYLTRANSFERASE SMYD2, N-[1-(N'-CYANO-N-[3-(DIFLUOROMETHOXY)PHENYL]CARBAMIMIDOYL)-3-(3,4-DICHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-4-YL]-N-ETHYL-2-HYDROXYACETAMIDE, ... | | Authors: | Hillig, R.C, Badock, V, Barak, N, Stellfeld, T, Eggert, E, ter Laak, A, Weiske, J, Christ, C.D, Koehr, S, Stoeckigt, D, Mowat, J, Mueller, T, Fernandez-Montalvan, A.E, Hartung, I.V, Stresemann, C, Brumby, T, Weinmann, H. | | Deposit date: | 2015-09-24 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Discovery and Characterization of a Highly Potent and Selective Aminopyrazoline-Based in Vivo Probe (Bay-598) for the Protein Lysine Methyltransferase Smyd2.
J.Med.Chem., 59, 2016
|
|
7TOC
 
 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | Authors: | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
