1BQO
 
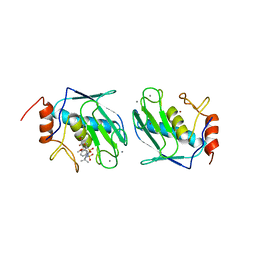 | | DISCOVERY OF POTENT, ACHIRAL MATRIX METALLOPROTEINASE INHIBITORS | | Descriptor: | 1,3-BIS-(4-METHOXY-BENZENESULFONYL)-5,5-DIMETHYL-HEXAHYDRO-PYRIMIDINE-2-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Pikul, S, Dunham, K.L.M, Almstead, N.G, De, B, Natchus, M.G, Anastasio, M.V, Mcphail, S.J, Snider, C.E, Taiwo, Y.O, Rydel, T.J, Dunaway, C.M, Gu, F, Mieling, G.E. | | Deposit date: | 1998-08-17 | | Release date: | 1999-08-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent, achiral matrix metalloproteinase inhibitors.
J.Med.Chem., 41, 1998
|
|
7OFZ
 
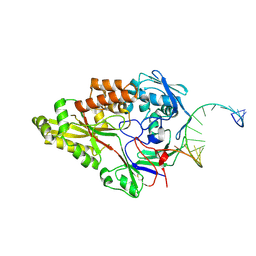 | | Nontypeable Haemophillus influenzae SapA in complex with double stranded RNA | | Descriptor: | ABC-type transport system, periplasmic component, involved in antimicrobial peptide resistance, ... | | Authors: | Lukacik, P, Owen, C.D, Nettleship, J.E, Bird, L.E, Owens, R.J, Walsh, M.A. | | Deposit date: | 2021-05-05 | | Release date: | 2021-10-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | The structure of nontypeable Haemophilus influenzae SapA in a closed conformation reveals a constricted ligand-binding cavity and a novel RNA binding motif.
Plos One, 16, 2021
|
|
6QRX
 
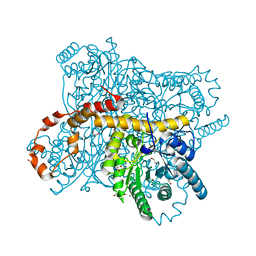 | | X-ray radiation dose series on xylose isomerase - 3.88 MGy | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Taberman, H, Bury, C.S, van der Woerd, M.J, Snell, E.H, Garman, E.F. | | Deposit date: | 2019-02-19 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Structural knowledge or X-ray damage? A case study on xylose isomerase illustrating both.
J.Synchrotron Radiat., 26, 2019
|
|
8P2W
 
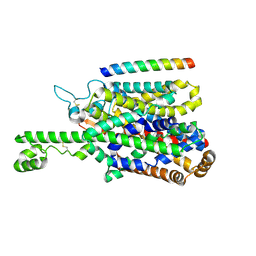 | | Structure of human SIT1 (focussed map / refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Sodium- and chloride-dependent transporter XTRP3 | | Authors: | Li, H.Z, Pike, A.C.W, Chi, G, Hansen, J.S, Lee, S.G, Rodstrom, K.E.J, Bushell, S.R, Speedman, D, Evans, A, Wang, D, He, D, Shrestha, L, Nasrallah, C, Chalk, R, Moreira, T, MacLean, E.M, Marsden, B, Bountra, C, Burgess-Brown, N.A, Dafforn, T.R, Carpenter, E.P, Sauer, D.B. | | Deposit date: | 2023-05-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structure and function of the SIT1 proline transporter in complex with the COVID-19 receptor ACE2.
Nat Commun, 15, 2024
|
|
8P2Q
 
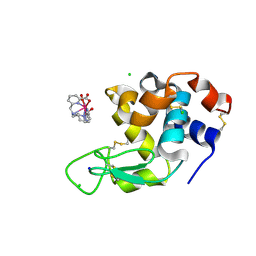 | |
6R0C
 
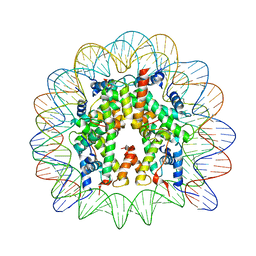 | | Human-D02 Nucleosome Core Particle with biotin-streptavidin label | | Descriptor: | DNA (142-MER), Histone H2A type 1, Histone H2B type 1-C/E/F/G/I, ... | | Authors: | Pye, V.E, Wilson, M.D, Cherepanov, P, Costa, A. | | Deposit date: | 2019-03-12 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
7ZBE
 
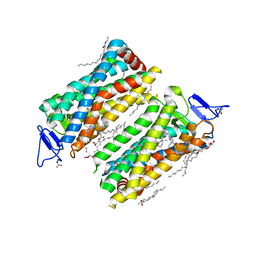 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SwissFEL) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, D, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
5OES
 
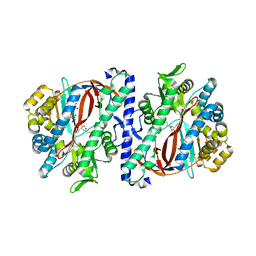 | | The structure of a glutathione synthetase (StGSS1) from Solanum tuberosum in ADP and y-EC bound closed conformation. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GAMMA-GLUTAMYLCYSTEINE, Glutathione synthetase, ... | | Authors: | Lilley, C.J, Maqbool, A, Wu, D, Yusup, H.B, Jones, L.M, Birch, P.R.J, Banfield, M.J, Urwin, P.E, Eves-van den Akker, S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-04-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Effector gene birth in plant parasitic nematodes: Neofunctionalization of a housekeeping glutathione synthetase gene.
PLoS Genet., 14, 2018
|
|
4YXW
 
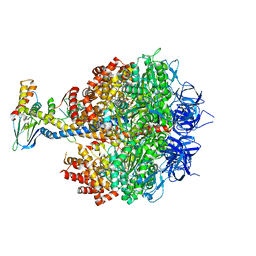 | | Bovine heart mitochondrial F1-ATPase inhibited by AMP-PNP and ADP in the presence of thiophosphate. | | Descriptor: | ATP synthase subunit alpha, mitochondrial, ATP synthase subunit beta, ... | | Authors: | Bason, J.V, Montgomery, M.G, Leslie, A.G.W, Walker, J.E. | | Deposit date: | 2015-03-23 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | How release of phosphate from mammalian F1-ATPase generates a rotary substep.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7S3F
 
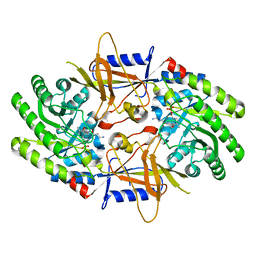 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
7S3G
 
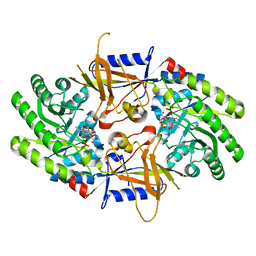 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with citrate at the catalytic center | | Descriptor: | CITRIC ACID, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
5OPT
 
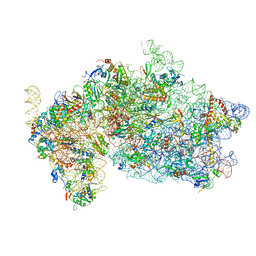 | | Structure of KSRP in context of Trypanosoma cruzi 40S | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, putative, ... | | Authors: | Brito Querido, J, Mancera-Martinez, E, Vicens, Q, Bochler, A, Chicher, J, Simonetti, A, Hashem, Y. | | Deposit date: | 2017-08-10 | | Release date: | 2017-11-15 | | Last modified: | 2017-12-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The cryo-EM Structure of a Novel 40S Kinetoplastid-Specific Ribosomal Protein.
Structure, 25, 2017
|
|
5VMY
 
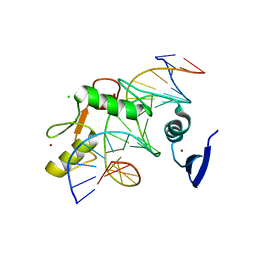 | | Kaiso (ZBTB33) zinc finger DNA binding domain in complex with a hemi CpG-methylated DNA resembling the specific Kaiso binding sequence (KBS) | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*CP*GP*CP*GP*GP*GP*AP*AP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*TP*TP*CP*CP*(5CM)P*GP*(5CM)P*GP*AP*AP*TP*AP*AP*CP*G)-3'), ... | | Authors: | Nikolova, E.N, Stanfield, R.L, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2017-04-28 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | CH···O Hydrogen Bonds Mediate Highly Specific Recognition of Methylated CpG Sites by the Zinc Finger Protein Kaiso.
Biochemistry, 57, 2018
|
|
7ZBC
 
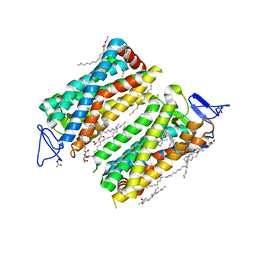 | | Dark state crystal structure of bovine rhodopsin in Lipidic Cubic Phase (SACLA) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gruhl, T, Weinert, T, Rodrigues, M.J, Milne, C, Ortolani, G, Nass, K, Nango, E, Sen, S, Johnson, P, Cirelli, C, Furrer, A, Mous, S, Skopintsev, P, James, D, Dworkowski, F, Baath, P, Kekilli, D, Oserov, D, Tanaka, R, Glover, H, Bacellar, C, Bruenle, S, Casadei, C, Diethelm, A, Gashi, D, Gotthard, G, Guixa-Gonzalez, R, Joti, Y, Kabanova, V, Knopp, G, Lesca, E, Ma, P, Martiel, I, Muehle, J, Owada, S, Pamula, F, Sarabi, S, Tejero, O, Tsai, C.J, Varma, N, Wach, A, Boutet, S, Tono, K, Nogly, P, Deupi, X, Iwata, S, Neutze, R, Standfuss, J, Schertler, G.F.X, Panneels, V. | | Deposit date: | 2022-03-23 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ultrafast structural changes direct the first molecular events of vision.
Nature, 615, 2023
|
|
6THN
 
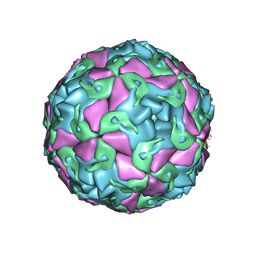 | | Multiple Genomic RNA-Coat Protein Contacts Play Vital Roles in the Assembly of Infectious Enterovirus-E symmetry expansion+2fold focused classification | | Descriptor: | Genome polyprotein, MYRISTIC ACID, RNA Peak 9 Bernoulli Plot, ... | | Authors: | Chandler-Bostock, R, Mata, C.P, Bingham, R, Dykeman, E.J, Meng, B, Tuthill, T.J, Rowlands, D.J, Ranson, N.A, Twarock, R, Stockley, P.G. | | Deposit date: | 2019-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly of infectious enteroviruses depends on multiple, conserved genomic RNA-coat protein contacts.
Plos Pathog., 16, 2020
|
|
1D4M
 
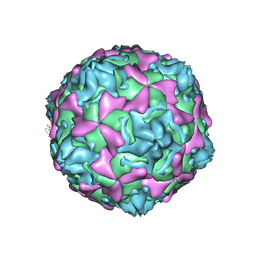 | | THE CRYSTAL STRUCTURE OF COXSACKIEVIRUS A9 TO 2.9 A RESOLUTION | | Descriptor: | 5-(7-(4-(4,5-DIHYDRO-2-OXAZOLYL)PHENOXY)HEPTYL)-3-METHYL ISOXAZOLE, MYRISTIC ACID, PROTEIN (COXSACKIEVIRUS A9) | | Authors: | Hendry, E, Hatanaka, H, Fry, E, Smyth, M, Tate, J, Stanway, G, Santti, J, Maaronen, M, Hyypia, T, Stuart, D. | | Deposit date: | 1999-10-04 | | Release date: | 1999-12-23 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses.
Structure Fold.Des., 7, 1999
|
|
4UHS
 
 | |
8DD2
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus Zolpidem | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Hibbs, R.E. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and dynamic mechanisms of GABA A receptor modulators with opposing activities.
Nat Commun, 13, 2022
|
|
8DD3
 
 | | Human GABAA receptor alpha1-beta2-gamma2 subtype in complex with GABA plus DMCM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Zhu, S, Hibbs, R.E. | | Deposit date: | 2022-06-17 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and dynamic mechanisms of GABA A receptor modulators with opposing activities.
Nat Commun, 13, 2022
|
|
6Z9W
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLGWVFAQV | | Descriptor: | Beta-2-microglobulin, LEU-LEU-GLY-TRP-VAL-PHE-ALA-GLN-VAL, MHC class I antigen | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
4UHK
 
 | |
8PUS
 
 | |
8QES
 
 | |
8Q7O
 
 | | Crystal structure of the FZD3 cysteine-rich domain in complex with a nanobody (14478) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Hillier, J.S, Zhao, Y, Malinauskas, T, Jones, E.Y. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural insights into Frizzled3 through nanobody modulators.
Nat Commun, 15, 2024
|
|
8AIE
 
 | | Crystal structure of D-amino acid aminotransferase from Aminobacterium colombiense complexed with D-cycloserine | | Descriptor: | 3-azanyloxy-2-[(~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylideneamino]propanoic acid, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Aminotransferase class IV, ... | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O, Bezsudnova, E.Y. | | Deposit date: | 2022-07-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 3D Structure of D-Аmino Acid Тransaminase from Aminobacterium colombiense in Complex with D-Cycloserine
Crystallography Reports, 68, 2023
|
|
