7SQD
 
 | |
7UVW
 
 | | A. baumannii ribosome: 70S with E-site tRNA | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.
Plos Biol., 21, 2023
|
|
7UVZ
 
 | | A. baumannii ribosome-Streptothricin-D complex: 70S with E-site tRNA | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.
Plos Biol., 21, 2023
|
|
7AKQ
 
 | |
6OZ6
 
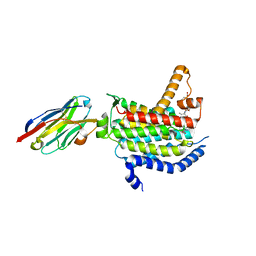 | | Crystal structure of MraY bound to 3'-hydroxymureidomycin A | | Descriptor: | (2~{S})-2-[[(2~{S})-1-[[(2~{S},3~{S})-3-[[(2~{S})-2-azanyl-3-(3-hydroxyphenyl)propanoyl]-methyl-amino]-1-[[(~{Z})-[(3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-ylidene]methyl]amino]-1-oxidanylidene-butan-2-yl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]carbamoylamino]-3-(3-hydroxyphenyl)propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
8TAR
 
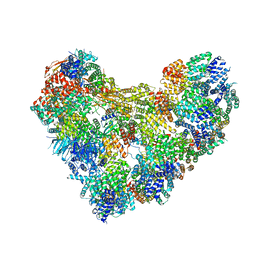 | | APC/C-CDH1-UBE2C-Ubiquitin-CyclinB-NTD | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
6PFZ
 
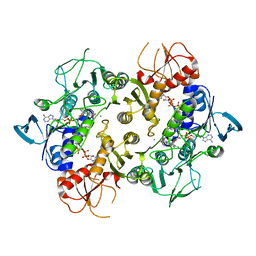 | | Structure of a NAD-Dependent Persulfide Reductase from A. fulgidus | | Descriptor: | CALCIUM ION, COENZYME A, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Sazinsky, M.H, Shabdar, S, Garcia-Constineiras, A, Crane III, E.J. | | Deposit date: | 2019-06-23 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.10003877 Å) | | Cite: | Structural and Kinetic Characterization of Hyperthermophilic NADH-Dependent Persulfide Reductase from Archaeoglobus fulgidus .
Archaea, 2021, 2021
|
|
7ALL
 
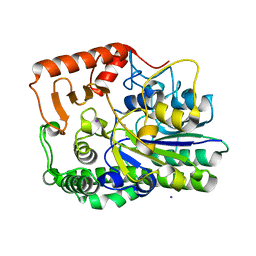 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT4683-S1_4) | | Descriptor: | Arylsulfatase, CALCIUM ION, IODIDE ION, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Martens, E.C, Basle, A, Cartmell, A. | | Deposit date: | 2020-10-06 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7YWY
 
 | |
7ANB
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-acetylgalactosamine-6-sulfatase, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-10-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7NYC
 
 | | cryoEM structure of 3C9-sMAC | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Menny, A, Couves, E.C, Bubeck, D. | | Deposit date: | 2021-03-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Structural basis of soluble membrane attack complex packaging for clearance.
Nat Commun, 12, 2021
|
|
8TAU
 
 | | APC/C-CDH1-UBE2C-UBE2S-Ubiquitin-CyclinB | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Bodrug, T, Welsh, K.A, Bolhuis, D.L, Paulakonis, E, Martinez-Chacin, R.C, Liu, B, Pinkin, N, Bonacci, T, Cui, L, Xu, P, Roscow, O, Amann, S.J, Grishkovskaya, I, Emanuele, M.J, Harrison, J.S, Steimel, J.P, Hahn, K.M, Zhang, W, Zhong, E, Haselbach, D, Brown, N.G. | | Deposit date: | 2023-06-27 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Time-resolved cryo-EM (TR-EM) analysis of substrate polyubiquitination by the RING E3 anaphase-promoting complex/cyclosome (APC/C).
Nat.Struct.Mol.Biol., 30, 2023
|
|
7ANA
 
 | | A single sulfatase is required for metabolism of colonic mucin O-glycans and intestinal colonization by a symbiotic human gut bacterium (BT1622-S1_20) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, ... | | Authors: | Sofia de Jesus Vaz Luis, A, Basle, A, Martens, E.C, Cartmell, A. | | Deposit date: | 2020-10-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A single sulfatase is required to access colonic mucin by a gut bacterium.
Nature, 598, 2021
|
|
7APD
 
 | | Bovine Papillomavirus E1 DNA helicase-replication fork complex | | Descriptor: | DNA (36-MER), DNA (40-MER), Replication protein E1 | | Authors: | Javed, A, Major, B, Stead, J, Sanders, C.M, Orlova, E.V. | | Deposit date: | 2020-10-16 | | Release date: | 2021-11-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Unwinding of a DNA replication fork by a hexameric viral helicase.
Nat Commun, 12, 2021
|
|
7YWH
 
 | | Six DNA Helix Bundle nanopore - State 1 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7YWO
 
 | | Six DNA Helix Bundle nanopore - State 5 | | Descriptor: | DNA (50-MER) | | Authors: | Javed, A, Ahmad, K, Lanphere, C, Coveney, P, Howorka, S, Orlova, E.V. | | Deposit date: | 2022-02-14 | | Release date: | 2023-05-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure and dynamics of an archetypal DNA nanoarchitecture revealed via cryo-EM and molecular dynamics simulations.
Nat Commun, 14, 2023
|
|
7B3I
 
 | | Notum complex with ARUK3003776 | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-chlorophenyl)sulfanyl-1$l^{4},2,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7B3H
 
 | | Notum complex with ARUK3003909 | | Descriptor: | 1,2-ETHANEDIOL, 6-((3-(trifluoromethoxy)phenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7B3P
 
 | | Notum complex with ARUK3003775 | | Descriptor: | 1,2-ETHANEDIOL, 6-((4-chlorophenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7B3X
 
 | | Notum complex with ARUK3003748 | | Descriptor: | 6-(m-tolylthio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Fish, P, Jones, E.Y. | | Deposit date: | 2020-12-01 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
6MZC
 
 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
7B45
 
 | | Notum complex with ARUK3003934 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-(3-methylsulfanylphenyl)sulfanyl-2~{H}-[1,2,4]triazolo[4,3-b]pyridazin-3-one, ... | | Authors: | Fish, P, Jones, E.Y. | | Deposit date: | 2020-12-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
4Y8J
 
 | | Yeast 20S proteasome in complex with Ac-LLL-ep | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Ac-PLL-ep, CHLORIDE ION, ... | | Authors: | Huber, E.M, Groll, M. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Systematic Analyses of Substrate Preferences of 20S Proteasomes Using Peptidic Epoxyketone Inhibitors.
J.Am.Chem.Soc., 137, 2015
|
|
7B50
 
 | | Notum complex with ARUK3003778 | | Descriptor: | 3-(3,4-dichlorophenyl)sulfanyl-1$l^{4},2,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4,7-tetraen-9-one, DIMETHYL SULFOXIDE, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-03 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
7B4X
 
 | | Notum complex with ARUK3002697 | | Descriptor: | 1,2-ETHANEDIOL, 6-(4-methylphenyl)sulfanyl-2~{H}-[1,2,4]triazolo[4,3-b]pyridazin-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Jones, E.Y, Fish, P. | | Deposit date: | 2020-12-02 | | Release date: | 2022-01-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural Analysis and Development of Notum Fragment Screening Hits.
Acs Chem Neurosci, 13, 2022
|
|
