1VC0
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Imidazole and Sr2+ solution | | Descriptor: | Hepatitis Delta virus ribozyme, STRONTIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VC6
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Product with C75U Mutaion, cleaved in Imidazole and Mg2+ solutions | | Descriptor: | Hepatitis Delta virus ribozyme, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VBX
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VC5
 
 | | Crystal Structure of the Wild Type Hepatitis Delta Virus Gemonic Ribozyme Precursor, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, SODIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VBZ
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Ba2+ solution | | Descriptor: | BARIUM ION, Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
5VGB
 
 | | Crystal structure of NmeCas9 HNH domain bound to anti-CRISPR AcrIIC1 | | Descriptor: | Anti-CRISPR protein (AcrIIC1), CRISPR-associated endonuclease Cas9, GLYCEROL, ... | | Authors: | Harrington, L.B, Doxzen, K.W, Ma, E, Knott, G.J, Kranzusch, P.J, Doudna, J.A. | | Deposit date: | 2017-04-10 | | Release date: | 2017-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | A Broad-Spectrum Inhibitor of CRISPR-Cas9.
Cell, 170, 2017
|
|
5VVL
 
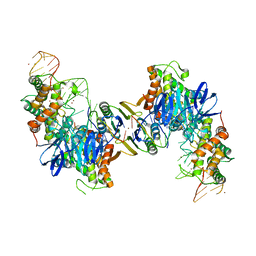 | | Cas1-Cas2 bound to full-site mimic with Ni | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (11-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5VZL
 
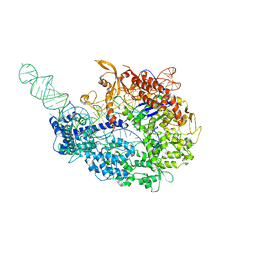 | | cryo-EM structure of the Cas9-sgRNA-AcrIIA4 anti-CRISPR complex | | Descriptor: | CRISPR-associated endonuclease Cas9, phage anti-CRISPR AcrIIA4, single guide RNA (116-MER) | | Authors: | Jiang, F, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-05-29 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Disabling Cas9 by an anti-CRISPR DNA mimic.
Sci Adv, 3, 2017
|
|
5UX0
 
 | |
5W1H
 
 | |
5VVK
 
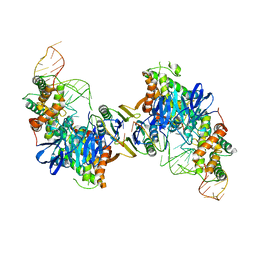 | | Cas1-Cas2 bound to full-site mimic | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (5'-D(*GP*AP*CP*CP*AP*CP*CP*AP*GP*TP*G)-3'), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.D, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5VVJ
 
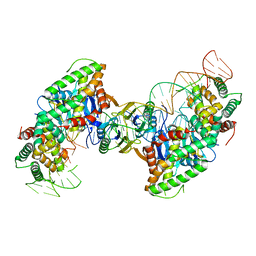 | | Cas1-Cas2 bound to half-site intermediate | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (112-MER), ... | | Authors: | Wright, A.V, Knott, G.J, Doxzen, K.W, Doudna, J.A. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5W1I
 
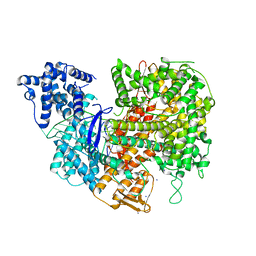 | |
5WFE
 
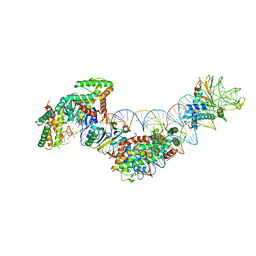 | | Cas1-Cas2-IHF-DNA holo-complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2, DNA (28-MER), ... | | Authors: | Wright, A.V, Liu, J.J, Nogales, E, Doudna, J.A. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structures of the CRISPR genome integration complex.
Science, 357, 2017
|
|
5WLH
 
 | |
4C9D
 
 | | Cas6 (TTHB231) product complex | | Descriptor: | CAS6B, R3 REPEAT RNA CLEAVAGE PRODUCT | | Authors: | Jinek, M, Niewoehner, O, Doudna, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases.
Nucleic Acids Res., 42, 2014
|
|
4C8Z
 
 | | Cas6 (TTHA0078) product complex | | Descriptor: | CAS6A, POTASSIUM ION, R1 REPEAT RNA CLEAVAGE PRODUCT, ... | | Authors: | Jinek, M, Niewoehner, O, Doudna, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Evolution of Crispr RNA Recognition and Processing by Cas6 Endonucleases.
Nucleic Acids Res., 42, 2014
|
|
4C8Y
 
 | |
4C98
 
 | |
4AN8
 
 | |
4C97
 
 | | Cas6 (TTHA0078) H37A mutant | | Descriptor: | CAS6A, SULFATE ION | | Authors: | Jinek, M, Niewoehner, O, Doudna, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of Crispr RNA Recognition and Processing by Cas6 Endonucleases.
Nucleic Acids Res., 42, 2014
|
|
4AL5
 
 | |
4AL6
 
 | |
4CMQ
 
 | | Crystal structure of Mn-bound S.pyogenes Cas9 | | Descriptor: | CRISPR-ASSOCIATED ENDONUCLEASE CAS9/CSN1, MANGANESE (II) ION, SULFATE ION | | Authors: | Jinek, M, Jiang, F, Taylor, D.W, Sternberg, S.H, Kaya, E, Ma, E, Anders, C, Hauer, M, Zhou, K, Lin, S, Kaplan, M, Iavarone, A.T, Charpentier, E, Nogales, E, Doudna, J.A. | | Deposit date: | 2014-01-17 | | Release date: | 2014-02-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of Cas9 Endonucleases Reveal RNA- Mediated Conformational Activation
Science, 343, 2014
|
|
4AL7
 
 | |
