1T7P
 
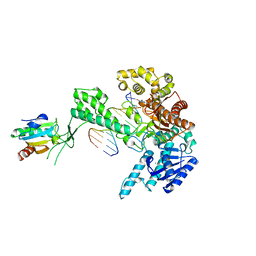 | | T7 DNA POLYMERASE COMPLEXED TO DNA PRIMER/TEMPLATE,A NUCLEOSIDE TRIPHOSPHATE, AND ITS PROCESSIVITY FACTOR THIOREDOXIN | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*CP*CP*TP*TP*GP*GP*CP*AP*CP*TP*GP*GP*C)-3'), DNA (5'-D(P*GP*CP*CP*AP*GP*TP*GP*CP*CP*AP*2DA)-3'), ... | | Authors: | Doublie, S, Tabor, S, Long, A.M, Richardson, C.C, Ellenberger, T. | | Deposit date: | 1997-09-24 | | Release date: | 1998-02-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution.
Nature, 391, 1998
|
|
1TDH
 
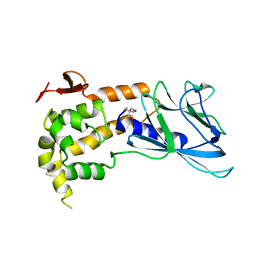 | | Crystal structure of human endonuclease VIII-like 1 (NEIL1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, nei endonuclease VIII-like 1 | | Authors: | Doublie, S, Bandaru, V, Bond, J.P, Wallace, S.S. | | Deposit date: | 2004-05-22 | | Release date: | 2004-07-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of human endonuclease VIII-like 1 (NEIL1) reveals a zincless finger motif required for glycosylase activity.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
4NRW
 
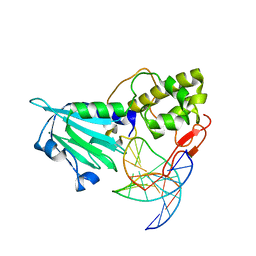 | | MvNei1-G86D | | Descriptor: | 5'-D(*CP*GP*TP*CP*CP*AP*(3DR)P*GP*TP*CP*TP*AP*C)-3', 5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3', formamidopyrimidine-DNA glycosylase | | Authors: | Prakash, A, Doublie, S. | | Deposit date: | 2013-11-27 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.845 Å) | | Cite: | Genome and cancer single nucleotide polymorphisms of the human NEIL1 DNA glycosylase: Activity, structure, and the effect of editing.
Dna Repair, 14, 2014
|
|
8SYJ
 
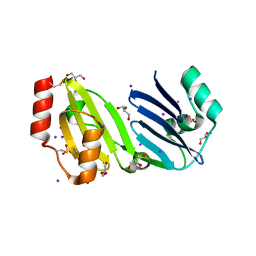 | | Structure of apurinic/apyrimidinic DNA lyase TK0353 from Thermococcus kodakarensis (Iodide crystal form) | | Descriptor: | GLYCEROL, IODIDE ION, SULFATE ION, ... | | Authors: | Eckenroth, B.E, Gardner, A.F, Doublie, S. | | Deposit date: | 2023-05-25 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermococcus kodakarensis TK0353 is a novel AP lyase with a new fold.
J.Biol.Chem., 300, 2023
|
|
8TH9
 
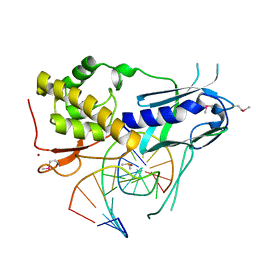 | |
9B15
 
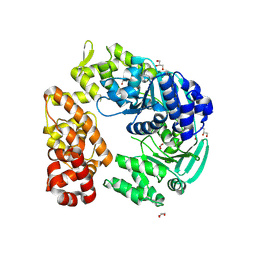 | |
4I0W
 
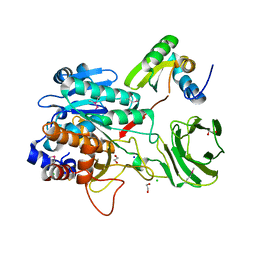 | | Structure of the Clostridium Perfringens CspB protease | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Protease CspB, ... | | Authors: | Adams, C.M, Eckenroth, B.E, Doublie, S. | | Deposit date: | 2012-11-19 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and functional analysis of the CspB protease required for Clostridium spore germination.
Plos Pathog., 9, 2013
|
|
4PPX
 
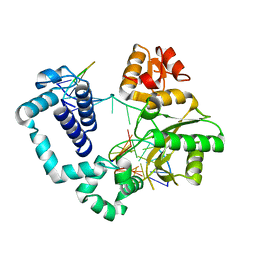 | | DNA Polymerase Beta E295K with Spiroiminodihydantoin in Templating Position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(SDH)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*G)-3', ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of DNA Polymerase beta with DNA Containing the Base Lesion Spiroiminodihydantoin in a Templating Position.
Biochemistry, 53, 2014
|
|
3L8B
 
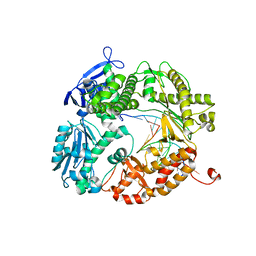 | | Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin | | Descriptor: | DNA (5'-D(*AP*C*TP*(G35)P*TP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*A)-3'), DNA polymerase, ... | | Authors: | Aller, P, Ye, Y, Wallace, S.S, Burrows, C.J, Doublie, S. | | Deposit date: | 2009-12-30 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a replicative DNA polymerase bound to the oxidized guanine lesion guanidinohydantoin.
Biochemistry, 49, 2010
|
|
3LDS
 
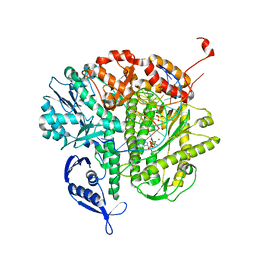 | | Crystal structure of RB69 gp43 with DNA and dATP opposite 8-oxoG | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*CP*AP*(8OG)P*CP*TP*TP*AP*TP*GP*AP*CP*AP*GP*CP*CP*GP*CP*G)-3'), DNA (5'-D(*GP*CP*GP*GP*CP*TP*GP*TP*CP*AP*TP*AP*AP*(DDG))-3'), ... | | Authors: | Hogg, M, Midkiff, J, Rudnicki, J, Reha-Krantz, L, Doublie, S, Wallace, S.S. | | Deposit date: | 2010-01-13 | | Release date: | 2010-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Kinetics of mismatch formation opposite lesions by the replicative DNA polymerase from bacteriophage RB69.
Biochemistry, 49, 2010
|
|
8GM7
 
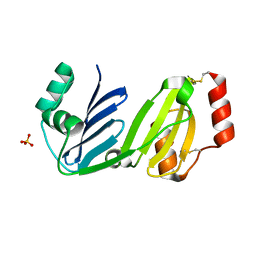 | |
8GM6
 
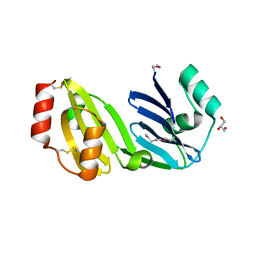 | |
6VJI
 
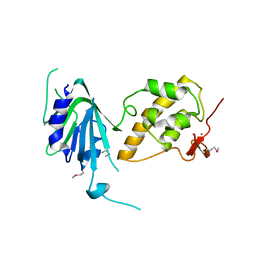 | |
7RAG
 
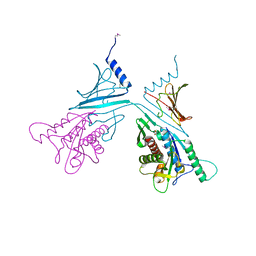 | |
7RDS
 
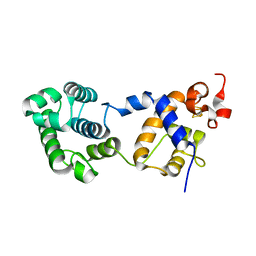 | | Structure of human NTHL1 | | Descriptor: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | Authors: | Carroll, B.L, Zahn, K.E, Doublie, S. | | Deposit date: | 2021-07-11 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
7RDT
 
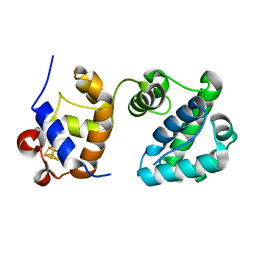 | | Structure of human NTHL1 - linker 1 chimera | | Descriptor: | IRON/SULFUR CLUSTER, Isoform 3 of Endonuclease III-like protein 1 | | Authors: | Carroll, B.L, Doublie, S. | | Deposit date: | 2021-07-11 | | Release date: | 2021-12-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Caught in motion: human NTHL1 undergoes interdomain rearrangement necessary for catalysis.
Nucleic Acids Res., 49, 2021
|
|
5U8H
 
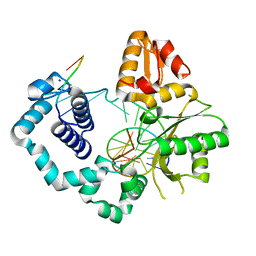 | | DNA Polymerase Beta G231D crystallized in PEG 400 | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.155 Å) | | Cite: | Remote Mutations Induce Functional Changes in Active Site Residues of Human DNA Polymerase beta.
Biochemistry, 56, 2017
|
|
6MW4
 
 | |
4X0P
 
 | |
4X0Q
 
 | |
3VK8
 
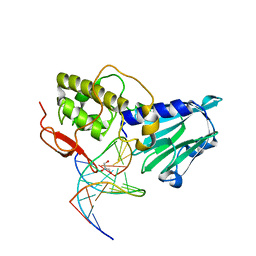 | | Crystal structure of DNA-glycosylase bound to DNA containing Thymine glycol | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(CTG)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*GP*AP*CP*CP*TP*GP*GP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Imamura, K, Averill, A, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-11-10 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA
J.Biol.Chem., 287, 2012
|
|
3VK7
 
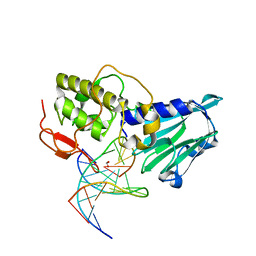 | | Crystal structure of DNA-glycosylase bound to DNA containing 5-Hydroxyuracil | | Descriptor: | DNA (5'-D(*CP*GP*TP*CP*CP*AP*(OHU)P*GP*TP*CP*TP*AP*C)-3'), DNA (5'-D(*GP*TP*AP*GP*AP*CP*GP*TP*GP*GP*AP*CP*G)-3'), FORMIC ACID, ... | | Authors: | Imamura, K, Averill, A, Wallace, S.S, Doublie, S. | | Deposit date: | 2011-11-10 | | Release date: | 2011-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of viral ortholog of human DNA glycosylase NEIL1 bound to thymine glycol or 5-hydroxyuracil-containing DNA
J.Biol.Chem., 287, 2012
|
|
5U8I
 
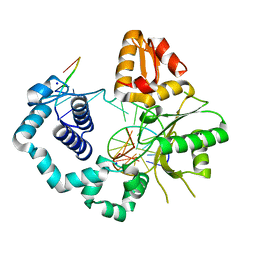 | | DNA Polymerase Beta S229L crystallized in PEG 400 | | Descriptor: | DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*TP*CP*GP*G)-3'), ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Remote Mutations Induce Functional Changes in Active Site Residues of Human DNA Polymerase beta.
Biochemistry, 56, 2017
|
|
5U8G
 
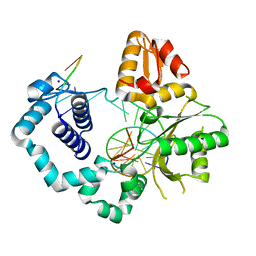 | | DNA Polymerase Beta crystallized in PEG 400 | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2016-12-14 | | Release date: | 2017-04-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.166 Å) | | Cite: | Remote Mutations Induce Functional Changes in Active Site Residues of Human DNA Polymerase beta.
Biochemistry, 56, 2017
|
|
1F5A
 
 | | CRYSTAL STRUCTURE OF MAMMALIAN POLY(A) POLYMERASE | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, POLY(A) POLYMERASE, ... | | Authors: | Martin, G, Keller, W, Doublie, S. | | Deposit date: | 2000-06-13 | | Release date: | 2000-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP.
EMBO J., 19, 2000
|
|
