7X5R
 
 | | Crystal structure of AtHPPD-Lancotrione complex | | Descriptor: | (1R,3R)-2-[(S)-[2-chloranyl-3-[2-(1,3-dioxolan-2-yl)ethoxy]-4-methylsulfonyl-phenyl]-oxidanyl-methyl]cyclohexane-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.644 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X62
 
 | | Crystal structure of AtHPPD-Y191051 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X6M
 
 | | Crystal structure of AtHPPD-Y191058 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, 5-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-2-(trifluoromethyl)quinazolin-4-one, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.815 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X5U
 
 | | Crystal structure of AtHPPD-diketonitrile complex | | Descriptor: | (1R,2S,3R)-1-cyclopropyl-2-(iminomethyl)-3-[2-methylsulfonyl-4-(trifluoromethyl)phenyl]propane-1,3-diol, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-hydroxyphenylpyruvate dioxygenase, ... | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-05 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
7X64
 
 | | Crystal structure of AtHPPD-Y18027 complex | | Descriptor: | 1,5-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-06 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Discovery of Subnanomolar Inhibitors of 4-Hydroxyphenylpyruvate Dioxygenase via Structure-Based Rational Design.
J.Agric.Food Chem., 71, 2023
|
|
7X6N
 
 | | Crystal structure of AtHPPD-Y191710 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, ethyl 5-methyl-4-oxidanylidene-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-phenyl-quinazoline-2-carboxylate | | Authors: | Lin, H.-Y, Dong, J, Yang, G.-F. | | Deposit date: | 2022-03-07 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.691 Å) | | Cite: | Structural insights of 4-Hydrophenylpyruvate dioxygenase inhibition by structurally diverse small molecules
Adv Agrochem, 2022
|
|
8WQM
 
 | | Complex structure of AtHPPD with Atovaquone | | Descriptor: | 2-[trans-4-(4-chlorophenyl)cyclohexyl]-3-hydroxynaphthalene-1,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-10-11 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Complex structure of AtHPPD with Atovaquone
To Be Published
|
|
8WI6
 
 | | Crystal structure of AtHPPD-ZSM824 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION, N-(1-methyl-1,2,3,4-tetrazol-5-yl)-1-pentyl-pyrrolo[2,3-b]pyridine-4-carboxamide | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-24 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.916 Å) | | Cite: | Crystal structure of AtHPPD-ZSM824 complex
To Be Published
|
|
8WHP
 
 | | Crystal structure of AtHPPD-YH201042 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-3-(2-ethoxyethyl)-5-methyl-2-(trifluoromethyl)quinazolin-4-one, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.919 Å) | | Cite: | Crystal structure of AtHPPD-YH201042 complex
To Be Published
|
|
8WHO
 
 | | Crystal structure of AtHPPD-YH20541 complex | | Descriptor: | 3-(ethylsulfanylmethyl)-~{N}-(5-methyl-1,3,4-oxadiazol-2-yl)-5-(trifluoromethyl)-[1,2,4]triazolo[4,3-a]pyridine-8-carboxamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | Crystal structure of AtHPPD-YH20541 complex
To Be Published
|
|
8WHQ
 
 | | Crystal structure of AtHPPD-YH21398 complex | | Descriptor: | 1,8-dimethyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-3-(phenylmethyl)quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-23 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Crystal structure of AtHPPD-YH21398 complex
To Be Published
|
|
8WK2
 
 | | Crystal structure of AtHPPD-F419Y+Y13287 complex | | Descriptor: | 1,5-dimethyl-3-(2-methylphenyl)-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.806 Å) | | Cite: | Crystal structure of AtHPPD-F419Y+Y13287 complex
To Be Published
|
|
8WL0
 
 | | Crystal structure of AtHPPD+Y19542 complex | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-(1,3-dimethyl-5-oxidanyl-pyrazol-4-yl)carbonyl-3-[(4-fluorophenyl)methyl]-5-methyl-1,2,3-benzotriazin-4-one, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-29 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.789 Å) | | Cite: | Crystal structure of AtHPPD-+Y19542 complex
To Be Published
|
|
8WJZ
 
 | | Crystal structure of AtHPPD-F419Y+Y13161 complex | | Descriptor: | 3-(2,6-dimethylphenyl)-1-methyl-6-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-09-26 | | Release date: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Crystal structure of AtHPPD-F419Y+Y13161 complex
To Be Published
|
|
8WPD
 
 | | Complex structure of AtHPPD with YH20009 | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, 6-chloranyl-3-(2,5-dimethylpyrazol-3-yl)-1-methyl-7-(2-oxidanyl-6-oxidanylidene-cyclohexen-1-yl)carbonyl-quinazoline-2,4-dione, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Complex structure of AtHPPD with YH20009
To Be Published
|
|
8WPC
 
 | | Complex structure of AtHPPD with LB553 | | Descriptor: | (2R)-3-[2,4-bis(chloranyl)-3-methyl-phenyl]carbonyl-1-cyclopropyl-2-methyl-4-oxidanyl-2H-pyrrol-5-one, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F, Lin, H.-Y, Dong, J. | | Deposit date: | 2023-10-09 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Complex structure of AtHPPD with LB553
To Be Published
|
|
6PCK
 
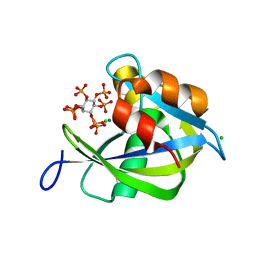 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 1-IP7 | | Descriptor: | (1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6PCL
 
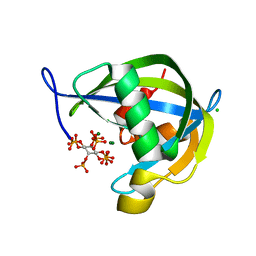 | | Crystal structure of human diphosphoinositol polyphosphate phosphohydrolase 1 in complex with 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Dollins, D.E, Neubauer, J, Dong, J, York, J.D. | | Deposit date: | 2019-06-17 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Vip1 is a kinase and pyrophosphatase switch that regulates inositol diphosphate signaling.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4HPW
 
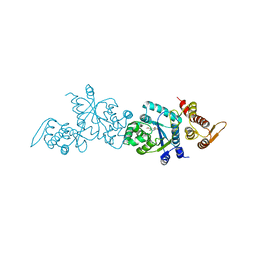 | | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine | | Descriptor: | 3-methoxy-L-tyrosine, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-24 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of Tyrosine-tRNA ligase mutant complexed with unnatural amino acid 3-o-methyl-Tyrosine
To be Published
|
|
4HK4
 
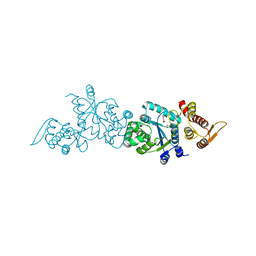 | | Crystal structure of apo Tyrosine-tRNA ligase mutant protein | | Descriptor: | DI(HYDROXYETHYL)ETHER, Tyrosine--tRNA ligase | | Authors: | Yu, Y, Zhou, Q, Dong, J, Li, J, Xiaoxuan, L, Mukherjee, A, Ouyang, H, Nilges, M, Li, H, Gao, F, Gong, W, Lu, Y, Wang, J. | | Deposit date: | 2012-10-15 | | Release date: | 2013-04-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Crystal structure of apo Tyrosine-tRNA ligase mutant protein
To be Published
|
|
4Q2V
 
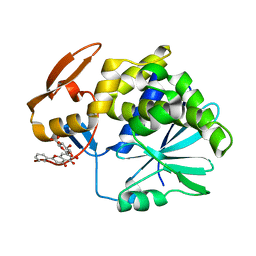 | | Crystal Structure of Ricin A chain complexed with Baicalin inhibitor | | Descriptor: | 5,6-dihydroxy-4-oxo-2-phenyl-4H-chromen-7-yl beta-D-glucopyranosiduronic acid, Ricin | | Authors: | Deng, X, Li, X, Dong, J, Chen, Y. | | Deposit date: | 2014-04-10 | | Release date: | 2015-04-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Baicalin inhibits the lethality of ricin in mice by inducing protein oligomerization.
J.Biol.Chem., 290, 2015
|
|
4OBY
 
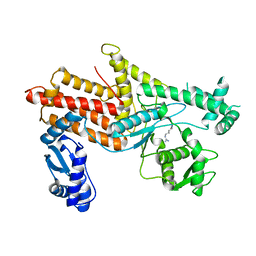 | | Crystal Structure of E.coli Arginyl-tRNA Synthetase and Ligand Binding Studies Revealed Key Residues in Arginine Recognition | | Descriptor: | ARGININE, Arginine--tRNA ligase | | Authors: | Bi, K, Zheng, Y, Dong, J, Gao, F, Wang, J, Wang, Y, Gong, W. | | Deposit date: | 2014-01-08 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.574 Å) | | Cite: | Crystal structure of E. coli arginyl-tRNA synthetase and ligand binding studies revealed key residues in arginine recognition.
Protein Cell, 5, 2014
|
|
4MYU
 
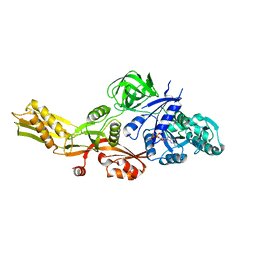 | |
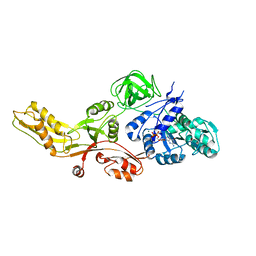
4MYT
 
 | |
5X5X
 
 | | Crystal structure of the Fab fragment of anti-osteocalcin C-terminal peptide antibody KTM219 | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, Fd chain of anti-osteocalcin antibody KTM219, ... | | Authors: | Komatsu, M, Dong, J, Ueda, H, Arai, R. | | Deposit date: | 2017-02-18 | | Release date: | 2018-02-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the Fab fragment of anti-osteocalcin C-terminal peptide antibody KTM219
To be published
|
|
