6ZM0
 
 | |
6I2V
 
 | | Pilotin from Vibrio vulnificus type 2 secretion system, EpsS. | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Howard, S.P, Estrozi, L, Bertrand, Q, Contreras-Martel, C, Strozen, T, Job, V, Martins, A, Fenel, D, Schoehn, G, Dessen, A. | | Deposit date: | 2018-11-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and assembly of pilotin-dependent and -independent secretins of the type II secretion system.
Plos Pathog., 15, 2019
|
|
4S3L
 
 | |
5A5F
 
 | | CRYSTAL STRUCTURE OF MURD LIGASE FROM ESCHERICHIA COLI IN COMPLEX WITH UMA AND ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MALONATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE, ... | | Authors: | Sink, R, Kotnik, M, Zega, A, Barreteau, H, Gobec, S, Blanot, D, Dessen, A, Contreras-Martel, C. | | Deposit date: | 2015-06-17 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic Study of Peptidoglycan Biosynthesis Enzyme MurD: Domain Movement Revisited.
PLoS ONE, 11, 2016
|
|
7BK8
 
 | | X-ray crystal structure of Pseudomonas aeruginosa MagC | | Descriptor: | MagC, PHOSPHATE ION | | Authors: | Zouhir, S, Contreras-Martel, C, Maragno Trindade, D, Attree, I, Dessen, A, Macheboeuf, P. | | Deposit date: | 2021-01-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | MagC is a NplC/P60-like member of the alpha-2-macroglobulin Mag complex of Pseudomonas aeruginosa that interacts with peptidoglycan.
Febs Lett., 595, 2021
|
|
3G66
 
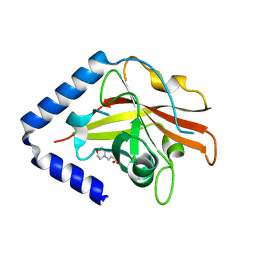 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
3G69
 
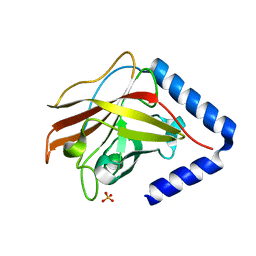 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
2C5W
 
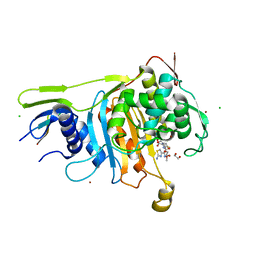 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) ACYL-ENZYME COMPLEX (CEFOTAXIME) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | 1,2-ETHANEDIOL, CEFOTAXIME, C3' cleaved, ... | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-02 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
2C6W
 
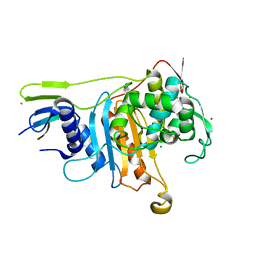 | | PENICILLIN-BINDING PROTEIN 1A (PBP-1A) FROM STREPTOCOCCUS PNEUMONIAE | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1A, ZINC ION | | Authors: | Contreras-Martel, C, Job, V, Di Guilmi, A.-M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2005-11-14 | | Release date: | 2005-12-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal Structure of Penicillin-Binding Protein 1A (Pbp1A) Reveals a Mutational Hotspot Implicated in Beta-Lactam Resistance in Streptococcus Pneumoniae.
J.Mol.Biol., 355, 2006
|
|
5FYA
 
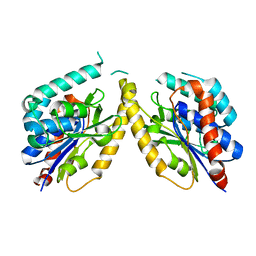 | | Cubic crystal of the native PlpD | | Descriptor: | PATATIN-LIKE PROTEIN, PLPD | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2016-03-04 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
5LP4
 
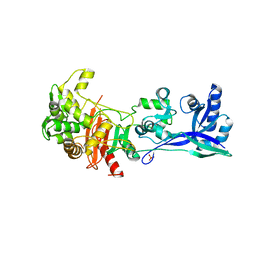 | | Penicillin-Binding Protein (PBP2) from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), SULFATE ION | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
5LP5
 
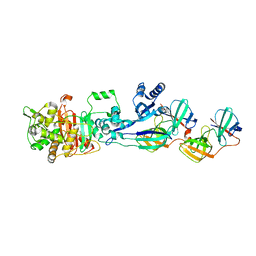 | | Complex between Penicillin-Binding Protein (PBP2) and MreC from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), Rod shape-determining protein (MreC) | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Hicham, S, Hardouin, P, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
5FQU
 
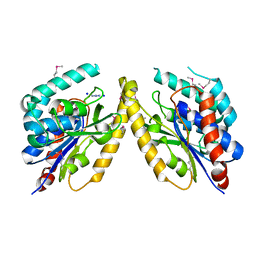 | | Orthorhombic crystal structure of of PlpD (selenomethionine derivative) | | Descriptor: | AZIDE ION, PATATIN-LIKE PROTEIN, PLPD, ... | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
1SBF
 
 | | SOYBEAN AGGLUTININ | | Descriptor: | CALCIUM ION, MANGANESE (II) ION, SOYBEAN AGGLUTININ, ... | | Authors: | Olsen, L.R, Dessen, A, Gupta, D, Sabesan, S, Brewer, C.F, Sacchettini, J.C. | | Deposit date: | 1997-10-21 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | X-ray crystallographic studies of unique cross-linked lattices between four isomeric biantennary oligosaccharides and soybean agglutinin.
Biochemistry, 36, 1997
|
|
2BG1
 
 | | Active site restructuring regulates ligand recognition in classA Penicillin-binding proteins (PBPs) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SULFATE ION | | Authors: | Macheboeuf, P, Di Guilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
1RP5
 
 | | PBP2x from Streptococcus pneumoniae strain 5259 with reduced susceptibility to beta-lactam antibiotics | | Descriptor: | SULFATE ION, penicillin-binding protein 2x | | Authors: | Pernot, L, Chesnel, L, Legouellec, A, Croize, J, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2003-12-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A PBP2x from a clinical isolate of Streptococcus pneumoniae exhibits an alternative mechanism for reduction of susceptibility to beta-lactam antibiotics.
J.Biol.Chem., 279, 2004
|
|
4BUC
 
 | | CRYSTAL STRUCTURE OF MURD LIGASE FROM THERMOTOGA MARITIMA IN APO FORM | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Favini-Stabile, S, Contreras-Martel, C, Thielens, N, Dessen, A. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Mreb and Murg as Scaffolds for the Cytoplasmic Steps of Peptidoglycan Biosynthesis
Environ.Microbiol., 15, 2013
|
|
4C12
 
 | | X-ray Crystal Structure of Staphylococcus aureus MurE with UDP-MurNAc- Ala-Glu-Lys and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Fulop, V, Roper, D.I, Ruane, K.M, Barreteau, H, Boniface, A, Dementin, S, Blanot, D, Mengin-Lecreulx, D, Gobec, S, Dessen, A, Dowson, C.G, Lloyd, A.J. | | Deposit date: | 2013-08-09 | | Release date: | 2013-10-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Specificity Determinants for Lysine Incorporation in Staphylococcus Aureus Peptidoglycan as Revealed by the Structure of a Mure Enzyme Ternary Complex.
J.Biol.Chem., 288, 2013
|
|
4CIH
 
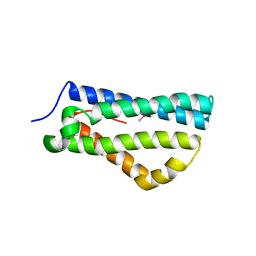 | | Structure of LntA-K180D-K181D from Listeria monocytogenes | | Descriptor: | LISTERIA NUCLEAR TARGETED PROTEIN A | | Authors: | Lebreton, A, Job, V, Ragon, M, Le Monnier, A, Dessen, A, Cossart, P, Bierne, H. | | Deposit date: | 2013-12-09 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural basis for the inhibition of the chromatin repressor BAHD1 by the bacterial nucleomodulin LntA.
MBio, 5, 2014
|
|
2JE5
 
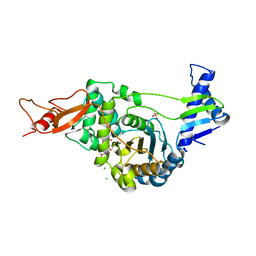 | | STRUCTURAL AND MECHANISTIC BASIS OF PENICILLIN BINDING PROTEIN INHIBITION BY LACTIVICINS | | Descriptor: | (2E)-2-{[(2S)-2-(ACETYLAMINO)-2-CARBOXYETHOXY]IMINO}PENTANEDIOIC ACID, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2007-01-15 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
4BUB
 
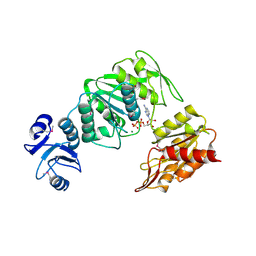 | | CRYSTAL STRUCTURE OF MURE LIGASE FROM THERMOTOGA MARITIMA IN COMPLEX WITH ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, UDP-N-ACETYLMURAMOYL-L-ALANYL-D-GLUTAMATE--LD-LYSINE LIGASE | | Authors: | Favini-Stabile, S, Contreras-Martel, C, Thielens, N, Dessen, A. | | Deposit date: | 2013-06-20 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mreb and Murg as Scaffolds for the Cytoplasmic Steps of Peptidoglycan Biosynthesis
Environ.Microbiol., 15, 2013
|
|
2JCH
 
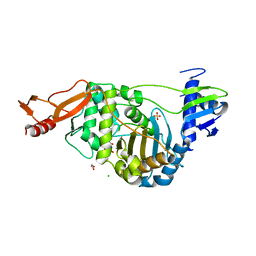 | | Structural and mechanistic basis of penicillin binding protein inhibition by lactivicins | | Descriptor: | (2E)-2-({(2S)-2-CARBOXY-2-[(PHENOXYACETYL)AMINO]ETHOXY}IMINO)PENTANEDIOIC ACID, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Macheboeuf, P, Fisher, D.S, Brown, T.J, Zervosen, A, Luxen, A, Joris, B, Dessen, A, Schofield, C.J. | | Deposit date: | 2006-12-23 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Mechanistic Basis of Penicillin-Binding Protein Inhibition by Lactivicins
Nat.Chem.Biol., 3, 2007
|
|
4A56
 
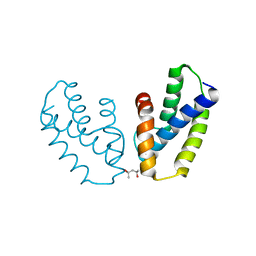 | | Crystal structure of the type 2 secretion system pilotin from Klebsiella Oxytoca | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, PULLULANASE SECRETION PROTEIN PULS | | Authors: | Tosi, T, Nickerson, N.N, Mollica, L, RingkjobingJensen, M, Blackledge, M, Baron, B, England, P, Pugsley, A.P, Dessen, A. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-07 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Pilotin-Secretin Recognition in the Type II Secretion System of Klebsiella Oxytoca.
Mol.Microbiol, 82, 2011
|
|
3ZM6
 
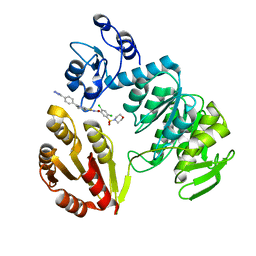 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | N-(6-(4-(2h-tetrazol-5-yl)benzyl)-3-cyano-4,5,6,7-tetrahydrothieno[2,3-c]pyridin-2-yl)-2,4-dichloro-5-(morpholinosulfonyl)benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
3ZM5
 
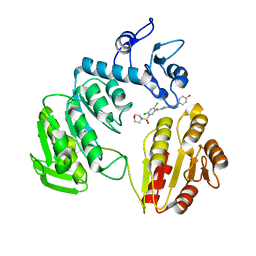 | | CRYSTAL STRUCTURE OF MURF LIGASE IN COMPLEX WITH CYANOTHIOPHENE INHIBITOR | | Descriptor: | 2,4-bis(chloranyl)-N-[3-cyano-6-[(4-hydroxyphenyl)methyl]-5,7-dihydro-4H-thieno[2,3-c]pyridin-2-yl]-5-morpholin-4-ylsulfonyl-benzamide, UDP-N-ACETYLMURAMOYL-TRIPEPTIDE--D-ALANYL-D-ALANINE LIGASE | | Authors: | Hrast, M, Turk, S, Sosic, I, Knez, D, Randall, C.P, Barreteau, H, Contreras-Martel, C, Dessen, A, ONeill, A.J, Mengin-Lecreulx, D, Blanot, D, Gobec, S. | | Deposit date: | 2013-02-05 | | Release date: | 2013-07-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure-Activity Relationships of New Cyanothiophene Inhibitors of the Essential Peptidoglycan Biosynthesis Enzyme Murf.
Eur.J.Med.Chem., 66C, 2013
|
|
