5AAC
 
 | | Structure of C1156Y Mutant Human Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2015-07-23 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
6AW4
 
 | | 1.50A resolution structure of catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catechol O-methyltransferase, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW9
 
 | | 2.55A resolution structure of SAH bound catechol O-methyltransferase (COMT) L136M from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW7
 
 | | 2.15A resolution structure of SAH bound catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW5
 
 | | 1.90A resolution structure of catechol O-methyltransferase (COMT) L136M (hexagonal form) from Nannospalax galili | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW8
 
 | | 2.25A resolution domain swapped dimer structure of SAH bound catechol O-methyltransferase (COMT) from Nannospalax galili | | Descriptor: | CALCIUM ION, Catechol O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
6AW6
 
 | | 1.70A resolution structure of catechol O-methyltransferase (COMT) L136M (rhombohedral form) from Nannospalax galili | | Descriptor: | Catechol O-methyltransferase, SULFATE ION | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Deng, Y, Hanzlik, R.P, Shams, I, Moskovitz, J. | | Deposit date: | 2017-09-05 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the catechol-o-methyl transferase (COMT) enzyme of the subterranean mole rat (Spalax) and the effect of L136M substitution
To be published
|
|
7SS1
 
 | | The structure of NTMT1 in complex with compound GD433 | | Descriptor: | (1R,3S,4R)-1-azabicyclo[2.2.2]octan-3-yl {2-[2-(4-fluoro-3-hydroxyphenyl)-1,3-thiazol-4-yl]propan-2-yl}carbamate, N-terminal Xaa-Pro-Lys N-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yadav, R, Guangping, D, Deng, Y, Huang, R, Noinaj, N. | | Deposit date: | 2021-11-09 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovery of a first-in-class small molecule inhibitor for Protein N-terminal methyltransferases 1/2
To Be Published
|
|
6B3W
 
 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-7-(3,5-dimethyl-1,2-oxazol-4-yl)-2-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit,Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
2YJR
 
 | |
2YHV
 
 | |
2X4Z
 
 | | Crystal Structure of the Human p21-Activated Kinase 4 in Complex with PF-03758309 | | Descriptor: | GLYCEROL, PF-3758309, SERINE/THREONINE-PROTEIN KINASE PAK 4 | | Authors: | Knighton, D.R, Deng, Y, Murray, B, Guo, C, Piraino, J, Westwick, J, Zhang, C, Lamerdin, J, Dagostino, E, Loi, C.-M, Zager, M, Kraynov, E, Christensen, J, Martinez, R, Kephart, S, Marakovits, J, Karlicek, S, Bergqvist, S, Smeal, T. | | Deposit date: | 2010-02-03 | | Release date: | 2010-05-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small-Molecule P21-Activated Kinase Inhibitor Pf- 3758309 is a Potent Inhibitor of Oncogenic Signaling and Tumor Growth.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2YFX
 
 | | Structure of L1196M Mutant Anaplastic Lymphoma Kinase in Complex with Crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, TYROSINE-PROTEIN KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A. | | Deposit date: | 2011-04-08 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design of Potent and Selective Inhibitors to Overcome Clinical Anaplastic Lymphoma Kinase Mutations Resistant to Crizotinib.
J.Med.Chem., 57, 2014
|
|
2YJS
 
 | |
2XP2
 
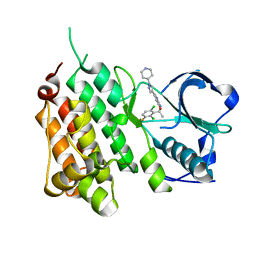 | | Structure of the Human Anaplastic Lymphoma Kinase in Complex with Crizotinib (PF-02341066) | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, TYROSINE-PROTEIN KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y, Liu, W, Brooun, A, Timofeevski, S, Marrone, T, Cui, J.J. | | Deposit date: | 2010-08-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure Based Drug Design of Crizotinib (Pf-02341066), a Potent and Selective Dual Inhibitor of Mesenchymal-Epithelial Transition Factor (C-met) Kinase and Anaplastic Lymphoma Kinase (Alk).
J.Med.Chem, 54, 2011
|
|
7CAI
 
 | | SARS-CoV-2 S trimer with two RBDs in the open state and complexed with two H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAH
 
 | | The interface of H014 Fab binds to SARS-CoV-2 S | | Descriptor: | Heavy chain of H014 Fab, Light chain of H014 Fab, Spike protein S1 | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Rao, Z, wang, Y, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-08-12 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAK
 
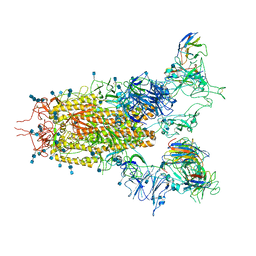 | | SARS-CoV-2 S trimer with three RBD in the open state and complexed with three H014 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-09-23 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAB
 
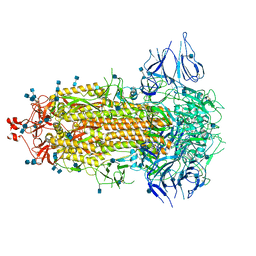 | | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7CAC
 
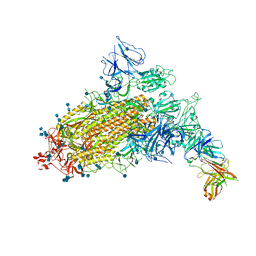 | | SARS-CoV-2 S trimer with one RBD in the open state and complexed with one H014 Fab. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of H014 Fab, ... | | Authors: | Zhe, L, Cao, L, Deng, Y, Sun, Y, Wang, N, Xie, L, Wang, Y, Rao, Z, Qin, C, Wang, X. | | Deposit date: | 2020-06-08 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody.
Science, 369, 2020
|
|
7W0J
 
 | | Acyl-CoA dehydrogenase, Tfu_1647 | | Descriptor: | Acyl-CoA dehydrogenase, [[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2R,3S,4S)-5-azanyl-2,3,4-tris(oxidanyl)pentyl] hydrogen phosphate | | Authors: | Liu, L.X, Zhou, S.H, Deng, Y. | | Deposit date: | 2021-11-18 | | Release date: | 2022-11-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Crystal structure of an Acyl-CoA dehydrogenase Tfu_1647
To Be Published
|
|
