3N2O
 
 | | X-ray crystal structure of arginine decarboxylase complexed with Arginine from Vibrio vulnificus | | Descriptor: | AGMATINE, Biosynthetic arginine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Deng, X, Lee, J, Michael, A.J, Tomchick, D.R, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2010-05-18 | | Release date: | 2010-06-09 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Evolution of substrate specificity within a diverse family of beta/alpha-barrel-fold basic amino acid decarboxylases: X-ray structure determination of enzymes with specificity for L-arginine and carboxynorspermidine.
J.Biol.Chem., 285, 2010
|
|
4IGH
 
 | | High resolution crystal structure of human dihydroorotate dehydrogenase bound with 4-quinoline carboxylic acid analog | | Descriptor: | 3-[decyl(dimethyl)ammonio]propane-1-sulfonate, 6-fluoro-2-[2-methyl-4-phenoxy-5-(propan-2-yl)phenyl]quinoline-4-carboxylic acid, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Deng, X, Das, P, Fontoura, B.M.A, Phillips, M.A, De Brabander, J.K. | | Deposit date: | 2012-12-17 | | Release date: | 2013-08-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | SAR Based Optimization of a 4-Quinoline Carboxylic Acid Analog with Potent Anti-Viral Activity.
ACS MED.CHEM.LETT., 4, 2013
|
|
4JPH
 
 | | Crystal structure of Protein Related to DAN and Cerberus (PRDC) | | Descriptor: | CITRIC ACID, GLUTATHIONE, GLYCEROL, ... | | Authors: | Deng, X, Nolan, K.T, Kattamuri, C, Thompson, T.B. | | Deposit date: | 2013-03-19 | | Release date: | 2013-07-24 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of protein related to dan and cerberus: insights into the mechanism of bone morphogenetic protein antagonism.
Structure, 21, 2013
|
|
4ORM
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM338 (N-[3,5-difluoro-4-(trifluoromethyl)phenyl]-5-methyl-2-(trifluoromethyl)[1,2,4]triazolo[1,5-a]pyrimidin-7-amine) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Fluorine Modulates Species Selectivity in the Triazolopyrimidine Class of Plasmodium falciparum Dihydroorotate Dehydrogenase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4OQV
 
 | | High resolution crystal structure of human dihydroorotate dehydrogenase bound with DSM338 (N-[3,5-difluoro-4-(trifluoromethyl)phenyl]-5-methyl-2-(trifluoromethyl)[1,2,4]triazolo[1,5-a]pyrimidin-7-amine) | | Descriptor: | ACETIC ACID, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2014-02-10 | | Release date: | 2014-06-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Fluorine Modulates Species Selectivity in the Triazolopyrimidine Class of Plasmodium falciparum Dihydroorotate Dehydrogenase Inhibitors.
J.Med.Chem., 57, 2014
|
|
4ORI
 
 | | Rat dihydroorotate dehydrogenase bound with DSM338 (N-[3,5-difluoro-4-(trifluoromethyl)phenyl]-5-methyl-2-(trifluoromethyl)[1,2,4]triazolo[1,5-a]pyrimidin-7-amine) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M.A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fluorine Modulates Species Selectivity in the Triazolopyrimidine Class of Plasmodium falciparum Dihydroorotate Dehydrogenase Inhibitors.
J.Med.Chem., 57, 2014
|
|
8JFU
 
 | | AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFO
 
 | |
8JFT
 
 | | Cryo-EM structure of SaCas9-AcrIIA15 CTD-sgRNA complex | | Descriptor: | AcrIIA15, CRISPR-associated endonuclease Cas9, sgRNA of SaCas9 | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JFR
 
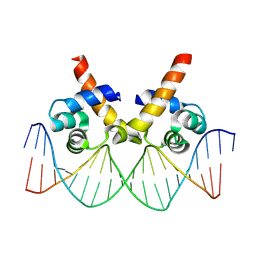 | | N-terminal domain of AcrIIA15 in complex with palindromic DNA substrate | | Descriptor: | AcrIIA15, DNA (5'-D(*AP*TP*TP*AP*TP*GP*AP*CP*AP*AP*AP*TP*GP*TP*CP*AP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*AP*TP*GP*AP*CP*AP*TP*TP*TP*GP*TP*CP*AP*TP*AP*A)-3') | | Authors: | Deng, X, Wang, Y. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | An anti-CRISPR that represses its own transcription while blocking Cas9-target DNA binding.
Nat Commun, 15, 2024
|
|
8JG9
 
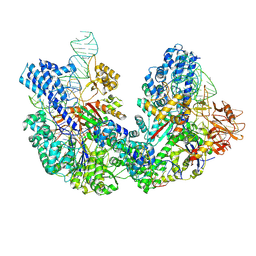 | |
7KH2
 
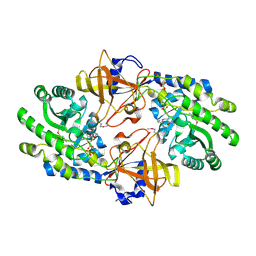 | | Structure of N-citrylornithine decarboxylase bound with PLP | | Descriptor: | GLYCEROL, N-citrylornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Deng, X, Tomchick, D, Phillips, M, Michael, A. | | Deposit date: | 2020-10-19 | | Release date: | 2020-12-16 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Alternative pathways utilize or circumvent putrescine for biosynthesis of putrescine-containing rhizoferrin.
J.Biol.Chem., 296, 2020
|
|
7L0K
 
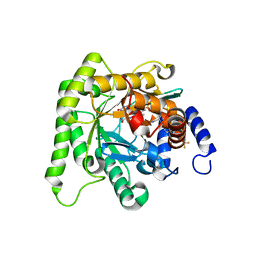 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM784 (3-(1-(3-methyl-4-((6-(trifluoromethyl)pyridin-3-yl)methyl)-1H-pyrrole-2-carboxamido)ethyl)-1H-pyrazole-5-carboxamide) | | Descriptor: | 3-{(1R)-1-[(3-methyl-4-{[6-(trifluoromethyl)pyridin-3-yl]methyl}-1H-pyrrole-2-carbonyl)amino]ethyl}-1H-pyrazole-5-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYK
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM589 (ethyl 3-methyl-4-((4-(trifluoromethyl)benzo[d]oxazol-7-yl)methyl)-1H-pyrrole-2-carboxylate) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZ4
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM705 (N-(1-(1H-1,2,4-triazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(1H-1,2,4-triazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYV
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM634 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(4-(trifluoromethyl)benzyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1,2-oxazol-3-yl)ethyl]-4-{[4-(trifluoromethyl)phenyl]methyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KZY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM778 (3-methyl-N-(1-(5-methyl-1H-pyrazol-3-yl)ethyl)-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1H-pyrazol-3-yl)ethyl]-4-{1-[6-(trifluoromethyl)pyridin-3-yl]cyclopropyl}-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7L01
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM782 (N-(1-(5-cyano-1H-pyrazol-3-yl)ethyl)-3-methyl-4-(1-(6-(trifluoromethyl)pyridin-3-yl)cyclopropyl)-1H-pyrrole-2-carboxamide) | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-10 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
7KYY
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM697 (3-methyl-N-(1-(5-methylisoxazol-3-yl)ethyl)-4-(6-(trifluoromethyl)-1H-indol-3-yl)-1H-pyrrole-2-carboxamide) | | Descriptor: | 3-methyl-N-[(1R)-1-(5-methyl-1,2-oxazol-3-yl)ethyl]-4-[6-(trifluoromethyl)-1H-indol-3-yl]-1H-pyrrole-2-carboxamide, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Deng, X, Phillips, M, Tomchick, D. | | Deposit date: | 2020-12-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent Antimalarials with Development Potential Identified by Structure-Guided Computational Optimization of a Pyrrole-Based Dihydroorotate Dehydrogenase Inhibitor Series.
J.Med.Chem., 64, 2021
|
|
5BOO
 
 | | Crystal structure of Plasmodium falciparum dihydroorotate dehydrogenase bound with Inhibitor DSM265 | | Descriptor: | 2-(1,1-difluoroethyl)-5-methyl-N-[4-(pentafluoro-lambda~6~-sulfanyl)phenyl][1,2,4]triazolo[1,5-a]pyrimidin-7-amine, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Phillips, M, Deng, X, Tomchick, D. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria.
Sci Transl Med, 7, 2015
|
|
5O01
 
 | | Crystal structure of BtubC (BKLC) from P. vanneervenii | | Descriptor: | BKLC (bacterial kinesin-light chain-like) | | Authors: | Lowe, J, Deng, X. | | Deposit date: | 2017-05-15 | | Release date: | 2017-07-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Four-stranded mini microtubules formed by Prosthecobacter BtubAB show dynamic instability.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CHH
 
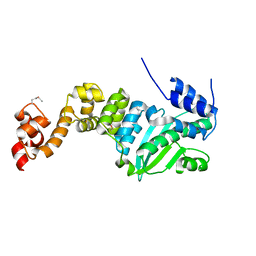 | | Crystal structure of transcriptional regulator CdpR from Pseudomonas aeruginosa | | Descriptor: | AraC family transcriptional regulator | | Authors: | Zhao, J.R, Yu, X, Zhu, M, Kang, H.P, Kong, W.N, Ma, J.B, Deng, X, Gan, J.H, Liang, H.H. | | Deposit date: | 2015-07-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Molecular Mechanism of CdpR Involved in Quorum-Sensing and Bacterial Virulence in Pseudomonas aeruginosa
Plos Biol., 14, 2016
|
|
8FTI
 
 | |
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
5E6M
 
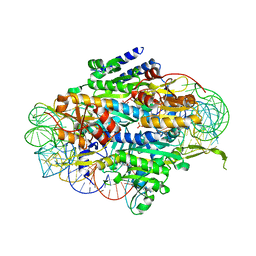 | | Crystal structure of human wild type GlyRS bound with tRNAGly | | Descriptor: | GLYCYL-ADENOSINE-5'-PHOSPHATE, Glycine--tRNA ligase, NICKEL (II) ION, ... | | Authors: | Xie, W, Qin, X, Deng, X, Chen, L, Liu, Y. | | Deposit date: | 2015-10-10 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.927 Å) | | Cite: | Crystal Structure of the Wild-Type Human GlyRS Bound with tRNA(Gly) in a Productive Conformation
J.Mol.Biol., 428, 2016
|
|
