8IVK
 
 | |
8HTY
 
 | | Candida boidinii Formate Dehydrogenase Crystal Structure at 1.4 Angstrom Resolution | | Descriptor: | Formate dehydrogenase, SULFATE ION | | Authors: | Gul, M, Yuksel, B, Bulut, H, DeMirci, H. | | Deposit date: | 2022-12-22 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural analysis of wild-type and Val120Thr mutant Candida boidinii formate dehydrogenase by X-ray crystallography.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8HZ2
 
 | |
8IQ7
 
 | |
8Z4B
 
 | |
8HZ1
 
 | |
8ZCM
 
 | | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
8ZCK
 
 | | Serial Femtosecond Crystallography Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Serial Femtosecond Crystallography Structure of Fc Fragment of Human IgG1 from VEGF-Trap
To Be Published
|
|
8ZCL
 
 | | Ambient Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ambient Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
8WZU
 
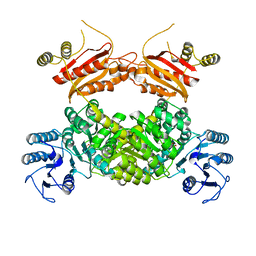 | |
8X34
 
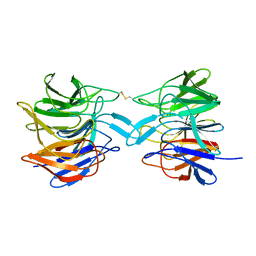 | |
8X6P
 
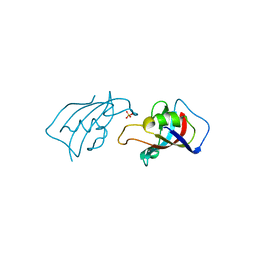 | | Isomerase Protein | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, SULFATE ION | | Authors: | Guven, O, Demirci, H. | | Deposit date: | 2023-11-21 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Isomerase Protein
To Be Published
|
|
6MPI
 
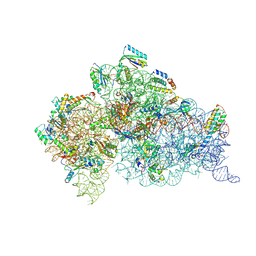 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-10-07 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
6MKN
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with an inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 2 (TRNAARG2) bound to an mRNA with an CGU-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-09-25 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
6MPF
 
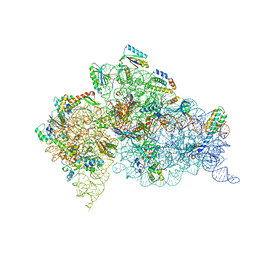 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGC-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-10-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
4GKK
 
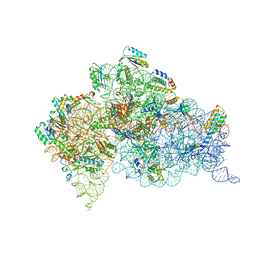 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human mitochondrial anticodon stem loop (ASL) of transfer RNA Methionine (TRNAMET) bound to an mRNA with an AUA-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, Murphy IV, F.V, Spears, J.L, Demirci, H, Agris, P.F. | | Deposit date: | 2012-08-11 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Expanded use of sense codons is regulated by modified cytidines in tRNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4GKJ
 
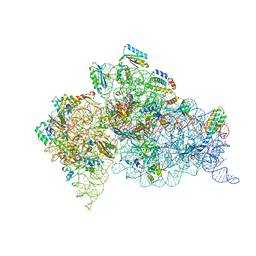 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human mitochondrial anticodon stem loop (ASL) of transfer RNA Methionine (TRNAMET) bound to an mRNA with an AUG-codon in the A-site and paromomycin. | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, Murphy IV, F.V, Spears, J.L, Demirci, H, Agris, P.F. | | Deposit date: | 2012-08-11 | | Release date: | 2013-06-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.298 Å) | | Cite: | Expanded use of sense codons is regulated by modified cytidines in tRNA.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
