8ORK
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | De Rose, S.A, Isupov, M. | | Deposit date: | 2023-04-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
8ORU
 
 | | cyclic 2,3-diphosphoglycerate synthetase from the hyperthermophilic archaeon Methanothermus fervidus bound to 2,3-diphosphoglycerate and ADP. | | Descriptor: | (2R)-2,3-diphosphoglyceric acid, 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | De Rose, S.A, Isupov, M. | | Deposit date: | 2023-04-17 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural characterization of a novel cyclic 2,3-diphosphoglycerate synthetase involved in extremolyte production in the archaeon Methanothermus fervidus .
Front Microbiol, 14, 2023
|
|
7NZQ
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-mannose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZO
 
 | | D-lyxose isomerasefrom the hyperthermophilic archaeon Thermofilum sp | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7NZP
 
 | | D-lyxose isomerase from the hyperthermophilic archaeon Thermofilum sp complexed with D-fructose | | Descriptor: | 1,2-ETHANEDIOL, D-lyxose/D-mannose family sugar isomerase, MANGANESE (II) ION, ... | | Authors: | De Rose, S.A, Isupov, M.N, Littlechild, J.A, Schoenheit, P. | | Deposit date: | 2021-03-24 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.345 Å) | | Cite: | Biochemical and Structural Characterisation of a Novel D-Lyxose Isomerase From the Hyperthermophilic Archaeon Thermofilum sp.
Front Bioeng Biotechnol, 9, 2021
|
|
7B0M
 
 | | Sugar transaminase from a metagenome collected from troll oil field production water | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Sugar aminotransferase, ... | | Authors: | Littlechild, J.A, De Rose, S.A, Isupov, M.N, Sayer, C, Karki, S. | | Deposit date: | 2020-11-20 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Sugar transaminases from hot environments
To Be Published
|
|
7B0D
 
 | | Sugar transaminase from Archaeoglobus veneficus | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | James, P, Littlechild, J.A, De Rose, S.A, Isupov, M.N. | | Deposit date: | 2020-11-19 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Sugar transaminases from hot environments
To Be Published
|
|
8AGS
 
 | | Cyclohexane epoxide soak of epoxide hydrolase from metagenomic source ch65 resulting in halogenated compound in the active site | | Descriptor: | 1,2-ETHANEDIOL, 2-CHLOROPHENOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8AGM
 
 | | Limonene epoxide low pH soak of epoxide hydrolase from metagenomic source ch65 | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, CHLORIDE ION, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.966 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8AGN
 
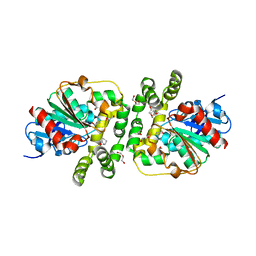 | | Cyclohexane epoxide low pH soak of epoxide hydrolase from metagenomic source ch65 | | Descriptor: | (1R,6S)-7-oxabicyclo[4.1.0]heptane, 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.957 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
8AGP
 
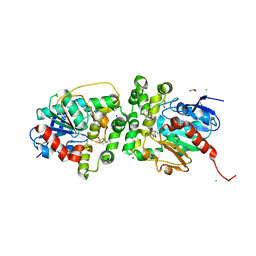 | | Halogenated product of limonene epoxide turnover by epoxide hydrolase from metagenomic source ch65 | | Descriptor: | (1~{S},2~{S},4~{R})-2-chloranyl-1-methyl-4-prop-1-en-2-yl-cyclohexan-1-ol, 1,2-ETHANEDIOL, Alpha/beta epoxide hydrolase, ... | | Authors: | Isupov, M.N, De Rose, S.A, Mitchell, D, Littlechild, J.A. | | Deposit date: | 2022-07-20 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Complexes of epoxide hydrolase from metagenomic source ch65
To Be Published
|
|
6T92
 
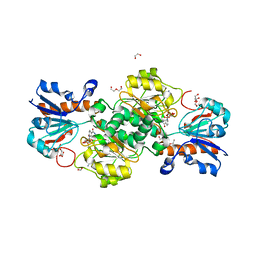 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Z
 
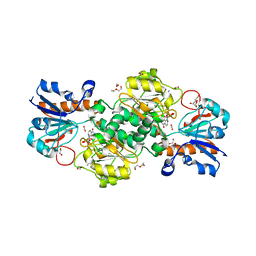 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A ternary complex with the oxidised form of the cofactor NAD+ and the substrate formate both at a primary and secondary sites. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T94
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex of N120C mutant protein with the reduced form of the cofactor NADH. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
6T8Y
 
 | | NAD+-dependent fungal formate dehydrogenase from Chaetomium thermophilum: A complex with the reduced form of the cofactor NADH and the substrate formate at a secondary site. | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Isupov, M.N, Yelmazer, B, De Rose, S.A, Littlechild, J.A. | | Deposit date: | 2019-10-25 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural insights into the NAD + -dependent formate dehydrogenase mechanism revealed from the NADH complex and the formate NAD + ternary complex of the Chaetomium thermophilum enzyme.
J.Struct.Biol., 212, 2020
|
|
5NFQ
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, IMIDAZOLE, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-15 | | Release date: | 2018-05-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
5NG7
 
 | | Novel epoxide hydrolases belonging to the alpha/beta hydrolases superfamily in metagenomes from hot environments | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ferrandi, E.E, De Rose, S.A, Sayer, C, Guazzelli, E, Marchesi, C, Saneei, V, Isupov, M.N, Littlechild, J.A, Monti, D. | | Deposit date: | 2017-03-17 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Thermophilic alpha / beta Class Epoxide Hydrolases Found in Metagenomes From Hot Environments.
Front Bioeng Biotechnol, 6, 2018
|
|
