6X7Z
 
 | | Inositol-bound structure of Marinomonas primoryensis PA14 carbohydrate-binding domain | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1,2-ETHANEDIOL, Antifreeze protein, ... | | Authors: | Guo, S, Davies, P.L. | | Deposit date: | 2020-06-01 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural Basis of Ligand Selectivity by a Bacterial Adhesin Lectin Involved in Multispecies Biofilm Formation.
Mbio, 12, 2021
|
|
6X9P
 
 | |
6X95
 
 | |
6X8A
 
 | |
6X8D
 
 | |
6X7T
 
 | |
6XA5
 
 | |
6X9M
 
 | |
6X7J
 
 | |
6X7Y
 
 | |
6X8Y
 
 | |
6XAC
 
 | |
6X6M
 
 | |
6X5V
 
 | |
6X5W
 
 | |
6X6Q
 
 | |
1N4I
 
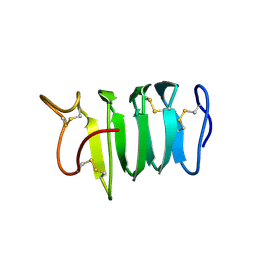 | | Solution structure of spruce budworm antifreeze protein at 5 degrees celsius | | Descriptor: | thermal hysteresis protein | | Authors: | Graether, S.P, Gagne, S.M, Spyracopoulos, L, Jia, Z, Davies, P.L, Sykes, B.D. | | Deposit date: | 2002-10-31 | | Release date: | 2003-04-08 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Spruce Budworm Antifreeze Protein: Changes in Structure and Dynamics at Low Temperature
J.Mol.Biol., 327, 2003
|
|
2G8J
 
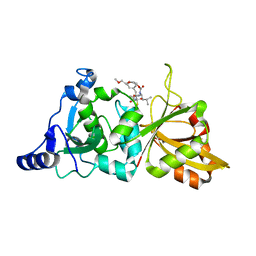 | | Calpain 1 proteolytic core in complex with SNJ-1945, a alpha-ketoamide-type inhibitor. | | Descriptor: | ((1S)-1-((((1S)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2,3-DIOXOPROPYL)AMINO)CARBONYL)-3-METHYLBUTYL)CARBAMIC ACID 5-METHOXY-3-OXAPENTYL ESTER, CALCIUM ION, Calpain-1 catalytic subunit | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
2G8E
 
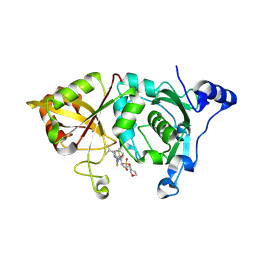 | | Calpain 1 proteolytic core in complex with SNJ-1715, a cyclic hemiacetal-type inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Calpain-1 catalytic subunit, ... | | Authors: | Cuerrier, D, Moldoveanu, T, Davies, P.L, Campbell, R.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-06-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Calpain Inhibition by alpha-Ketoamide and Cyclic Hemiacetal Inhibitors Revealed by X-ray Crystallography
Biochemistry, 45, 2006
|
|
5MSI
 
 | |
1EZG
 
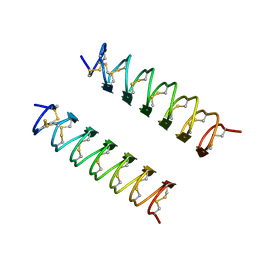 | | CRYSTAL STRUCTURE OF ANTIFREEZE PROTEIN FROM THE BEETLE, TENEBRIO MOLITOR | | Descriptor: | THERMAL HYSTERESIS PROTEIN ISOFORM YL-1 | | Authors: | Liou, Y.-C, Tocilj, A, Davies, P.L, Jia, Z. | | Deposit date: | 2000-05-10 | | Release date: | 2000-08-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mimicry of ice structure by surface hydroxyls and water of a beta-helix antifreeze protein.
Nature, 406, 2000
|
|
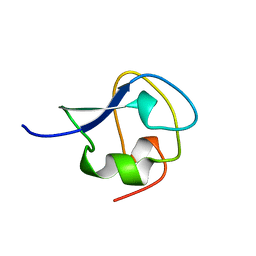
4MSI
 
 | |
4OKH
 
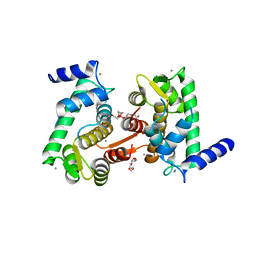 | | Crystal structure of calpain-3 penta-EF-hand domain | | Descriptor: | 2,5,8,11,14,17,20,23,26,29,32,35,38,41,44,47,50,53,56,59,62,65,68,71,74,77,80-HEPTACOSAOXADOOCTACONTAN-82-OL, CALCIUM ION, Calpain-3 | | Authors: | Karunan Partha, S, Ravulapalli, R, Campbell, R.L, Allingham, J.S, Davies, P.L. | | Deposit date: | 2014-01-22 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of calpain-3 penta-EF-hand (PEF) domain - a homodimerized PEF family member with calcium bound at the fifth EF-hand.
Febs J., 281, 2014
|
|
4P99
 
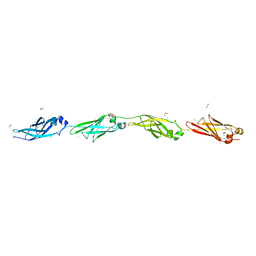 | | Ca2+-stabilized adhesin helps an Antarctic bacterium reach out and bind ice | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Guo, S, Vance, D.R.T, Campbell, R.L, Davies, P.L. | | Deposit date: | 2014-04-02 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ca2+-stabilized adhesin helps an Antarctic bacterium reach out and bind ice.
Biosci.Rep., 34, 2014
|
|
2NQI
 
 | | Calpain 1 proteolytic core inactivated by WR13(R,R), an epoxysuccinyl-type inhibitor. | | Descriptor: | CALCIUM ION, Calpain-1 catalytic subunit, N~2~-[(2S)-2-{[(2R)-4-ETHOXY-2-HYDROXY-4-OXOBUTANOYL]AMINO}PENT-4-ENOYL]-L-ARGINYL-L-TRYPTOPHANAMIDE | | Authors: | Cuerrier, D, Davies, P.L, Campbell, R.L, Moldoveanu, T. | | Deposit date: | 2006-10-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Development of Calpain-specific Inactivators by Screening of Positional Scanning Epoxide Libraries
J.Biol.Chem., 282, 2007
|
|
