2LXP
 
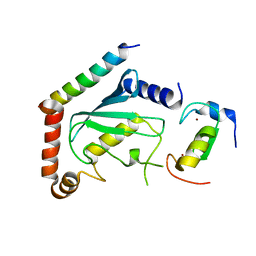 | | NMR structure of two domains in ubiquitin ligase gp78, RING and G2BR, bound to its conjugating enzyme Ube2g | | Descriptor: | E3 ubiquitin-protein ligase AMFR, Ubiquitin-conjugating enzyme E2 G2, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-30 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
2LXH
 
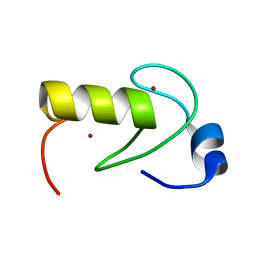 | | NMR structure of the RING domain in ubiquitin ligase gp78 | | Descriptor: | E3 ubiquitin-protein ligase AMFR, ZINC ION | | Authors: | Das, R, Linag, Y, Mariano, J, Li, J, Huang, T, King, A, Weissman, A, Ji, X, Byrd, R. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine.
Embo J., 32, 2013
|
|
8Q6J
 
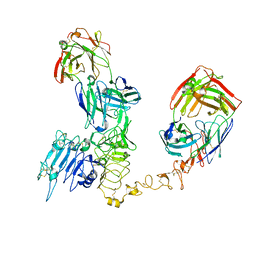 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-08-11 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
2JR0
 
 | | Solution structure of NusB from Aquifex Aeolicus | | Descriptor: | N utilization substance protein B homolog | | Authors: | Das, R, Loss, S, Li, J, Tarasov, S, Wingfield, P, Waugh, D.S, Byrd, R.A, Altieri, A.S. | | Deposit date: | 2007-06-18 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structural biophysics of the NusB:NusE antitermination complex.
J.Mol.Biol., 376, 2008
|
|
9EPT
 
 | |
9GP8
 
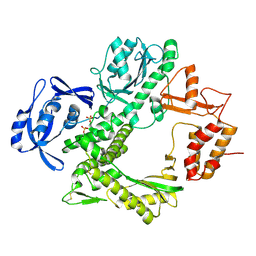 | |
8PWH
 
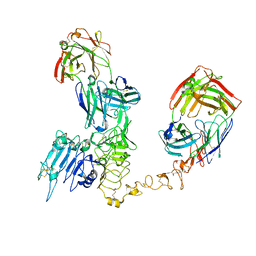 | | Atomic structure and conformational variability of the HER2-Trastuzumab-Pertuzumab complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Pertuzumab Fab heavy chain, Pertuzumab Fab light chain, ... | | Authors: | Ruedas, R, Vuillemot, R, Tubiana, T, Winter, J.M, Pieri, L, Arteni, A.A, Samson, C, Jonic, J, Mathieu, M, Bressanelli, S. | | Deposit date: | 2023-07-20 | | Release date: | 2024-02-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structure and conformational variability of the HER2-trastuzumab-pertuzumab complex.
J.Struct.Biol., 216, 2024
|
|
1V05
 
 | |
2BP3
 
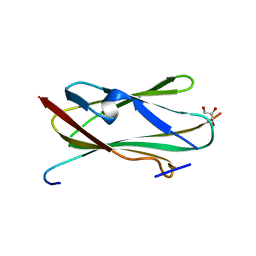 | |
2LFY
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dA | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
2LFX
 
 | | Structure of the duplex when (5'S)-8,5'-cyclo-2'-deoxyguanosine is placed opposite dT | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*TP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures of (5'S)-8,5'-Cyclo-2'-deoxyguanosine Mismatched with dA or dT.
Chem.Res.Toxicol., 25, 2012
|
|
2LG0
 
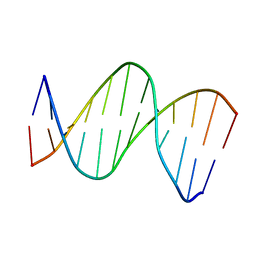 | | structure of the duplex containing (5'S)-8,5'-cyclo-2'-deoxyadenosine | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*TP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(02I)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-07-18 | | Release date: | 2012-06-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of (5'S)-8,5'-cyclo-2'-deoxyguanosine in DNA.
J.Am.Chem.Soc., 133, 2011
|
|
2LFA
 
 | | Oligonucleotide duplex contaning (5'S)-8,5'-cyclo-2'-deoxyguansine | | Descriptor: | DNA (5'-D(*AP*CP*AP*AP*AP*CP*AP*CP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*(2LF)P*TP*GP*TP*TP*TP*GP*T)-3') | | Authors: | Huang, H, Das, R.S, Basu, A, Stone, M.P. | | Deposit date: | 2011-06-29 | | Release date: | 2012-01-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of (5'S)-8,5'-Cyclo-2'-deoxyguanosine in DNA.
J.Am.Chem.Soc., 133, 2011
|
|
5XFU
 
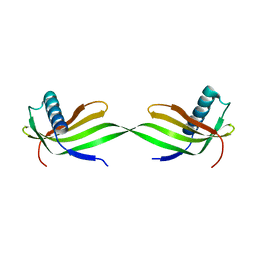 | | Domain swapped dimer crystal structure of loop1 deletion mutant in Single-chain Monellin | | Descriptor: | Monellin chain B,Monellin chain A | | Authors: | Surana, P, Nandwani, N, Udgaonkar, J, Gosavi, S, Das, R. | | Deposit date: | 2017-04-11 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.611 Å) | | Cite: | Amino-acid composition after loop deletion drives domain swapping
Protein Sci., 26, 2017
|
|
3GT8
 
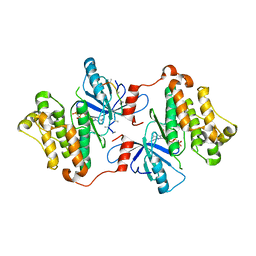 | | Crystal structure of the inactive EGFR kinase domain in complex with AMP-PNP | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Jura, N, Endres, N.F, Engel, K, Deindl, S, Das, R, Lamers, M.H, Wemmer, D.E, Zhang, X, Kuriyan, J. | | Deposit date: | 2009-03-27 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.955 Å) | | Cite: | Mechanism for activation of the EGF receptor catalytic domain by the juxtamembrane segment.
Cell(Cambridge,Mass.), 137, 2009
|
|
5XQM
 
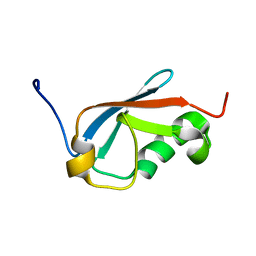 | |
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
9CBX
 
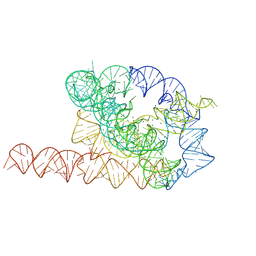 | | Tetrahymena ribozyme with automatically identified water and magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (387-MER) | | Authors: | Kretsch, R.C, Li, S, Pintilie, G, Palo, M.Z, Case, D.A, Das, R, Zhang, K, Chiu, W. | | Deposit date: | 2024-06-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Complex water networks visualized by cryogenic electron microscopy of RNA.
Nature, 2025
|
|
9CBY
 
 | | Tetrahymena ribozyme with automatically identified water and magnesium ions | | Descriptor: | MAGNESIUM ION, RNA (387-MER) | | Authors: | Kretsch, R.C, Li, S, Pintilie, G, Palo, M.Z, Case, D.A, Das, R, Zhang, K, Chiu, W. | | Deposit date: | 2024-06-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Complex water networks visualized by cryogenic electron microscopy of RNA.
Nature, 2025
|
|
6WLN
 
 | | hc16 ligase product models, 10.0 Angstrom resolution | | Descriptor: | RNA (349-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLL
 
 | | Apo F. nucleatum glycine riboswitch models, 10.0 Angstrom resolution | | Descriptor: | RNA (171-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
6WLJ
 
 | | ATP-TTR-3 with AMP models, 9.6 Angstrom resolution | | Descriptor: | RNA (130-MER) | | Authors: | Kappel, K, Zhang, K, Su, Z, Watkins, A.M, Kladwang, W, Li, S, Pintilie, G, Topkar, V.V, Rangan, R, Zheludev, I.N, Yesselman, J.D, Chiu, W, Das, R. | | Deposit date: | 2020-04-20 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Accelerated cryo-EM-guided determination of three-dimensional RNA-only structures.
Nat.Methods, 17, 2020
|
|
