3RZV
 
 | |
4JPX
 
 | |
3IRT
 
 | |
4JPY
 
 | |
3RZU
 
 | | The Crystal Structure of the Catalytic Domain of AMSH | | Descriptor: | STAM-binding protein, ZINC ION | | Authors: | Davies, C.W, Das, C. | | Deposit date: | 2011-05-12 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Thermodynamic Comparison of the Catalytic Domain of AMSH and AMSH-LP: Nearly Identical Fold but Different Stability.
J.Mol.Biol., 413, 2011
|
|
4JXE
 
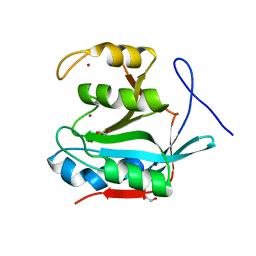 | | Crystal structure of Schizosaccharomyces pombe sst2 catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMSH-like protease sst2, ... | | Authors: | Shrestha, R.K, Das, C. | | Deposit date: | 2013-03-28 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
4JKJ
 
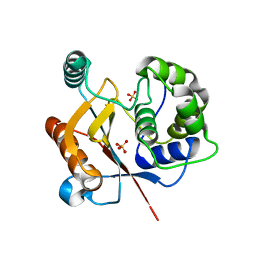 | |
4ETL
 
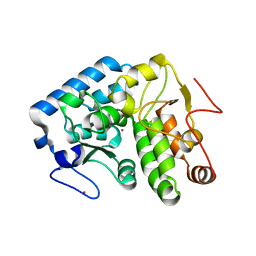 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum F258A mutation | | Descriptor: | COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Paul, L.P, Corn, I.R, Wagner, K.T, Abu-Omar, M.M, Das, C. | | Deposit date: | 2012-04-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
4ESM
 
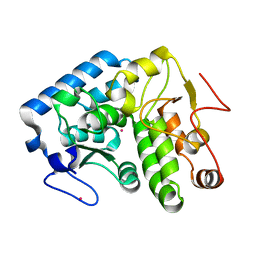 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum Y155A mutation | | Descriptor: | COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Paul, L.P, Corn, I.R, Wagner, K.T, Abu-Omar, M.M, Das, C. | | Deposit date: | 2012-04-23 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
4K1R
 
 | |
4DM9
 
 | | The Crystal Structure of Ubiquitin Carboxy-terminal hydrolase L1 (UCHL1) bound to a tripeptide fluoromethyl ketone Z-VAE(OMe)-FMK | | Descriptor: | Tripeptide fluoromethyl ketone inhibitor Z-VAE(OMe)-FMK, Ubiquitin carboxyl-terminal hydrolase isozyme L1 | | Authors: | Davies, C.W, Chaney, J, Korbel, G, Ringe, D, Petsko, G.A, Ploegh, H, Das, C. | | Deposit date: | 2012-02-07 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The co-crystal structure of ubiquitin carboxy-terminal hydrolase L1 (UCHL1) with a tripeptide fluoromethyl ketone (Z-VAE(OMe)-FMK).
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4I6N
 
 | | Crystal structure of Trichinella spiralis UCH37 catalytic domain bound to Ubiquitin vinyl methyl ester | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, METHYL 4-AMINOBUTANOATE, SODIUM ION, ... | | Authors: | Morrow, M.E, Ronau, J.A, White, R.R, Artavanis-Tsakonas, K, Das, C. | | Deposit date: | 2012-11-29 | | Release date: | 2013-05-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Stabilization of an Unusual Salt Bridge in Ubiquitin by the Extra C-Terminal Domain of the Proteasome-Associated Deubiquitinase UCH37 as a Mechanism of Its Exo Specificity.
Biochemistry, 52, 2013
|
|
4OUF
 
 | | Crystal Structure of CBP bromodomain | | Descriptor: | 1,2-ETHANEDIOL, CREB-binding protein, DI(HYDROXYETHYL)ETHER | | Authors: | Roy, S, Das, C, Tyler, J.K, Kutateladze, T.G. | | Deposit date: | 2014-02-17 | | Release date: | 2014-03-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4Q3Y
 
 | |
4Q3Z
 
 | |
4Q3W
 
 | | Crystal structure of C. violaceum phenylalanine hydroxylase D139E mutation | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, Phenylalanine-4-hydroxylase | | Authors: | Ronau, J.A, Abu-Omar, M.M, Das, C. | | Deposit date: | 2014-04-12 | | Release date: | 2015-02-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conserved acidic residue in phenylalanine hydroxylase contributes to cofactor affinity and catalysis.
Biochemistry, 53, 2014
|
|
4Q3X
 
 | |
4MS7
 
 | | Crystal structure of Schizosaccharomyces pombe sst2 catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMSH-like protease sst2, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
4PQT
 
 | | Insights into the mechanism of deubiquitination by JAMM deubiquitinases from co-crystal structures of enzyme with substrate and product | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease sst2, Protein UBBP4, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2014-03-04 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
4NQL
 
 | | The crystal structure of the DUB domain of AMSH orthologue, Sst2 from S. pombe, in complex with lysine 63-linked diubiquitin | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease sst2, Ubiquitin, ... | | Authors: | Ronau, J.A, Shrestha, R.K, Das, C. | | Deposit date: | 2013-11-25 | | Release date: | 2014-10-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into the mechanism of deubiquitination by JAMM deubiquitinases from cocrystal structures of the enzyme with the substrate and product.
Biochemistry, 53, 2014
|
|
4MSJ
 
 | | Crystal structure of S. pombe AMSH-like protease SST2 catalytic domain from P212121 space group | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease sst2, GLYCINE, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
4MSM
 
 | | Crystal structure of Schizosaccharomyces pombe AMSH-like protease sst2 E286A mutant bound to ubiquitin | | Descriptor: | 1,2-ETHANEDIOL, AMSH-like protease sst2, PHOSPHATE ION, ... | | Authors: | Shrestha, R.K, Ronau, J.A, Das, C. | | Deposit date: | 2013-09-18 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Insights into the Mechanism of Deubiquitination by JAMM Deubiquitinases from Cocrystal Structures of the Enzyme with the Substrate and Product.
Biochemistry, 53, 2014
|
|
3TCY
 
 | | Crystallographic structure of phenylalanine hydroxylase from Chromobacterium violaceum (cPAH) bound to phenylalanine in a site distal to the active site | | Descriptor: | 1,2-ETHANEDIOL, COBALT (II) ION, PHENYLALANINE, ... | | Authors: | Ronau, J.A, Abu-Omar, M.M, Das, C. | | Deposit date: | 2011-08-09 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An additional substrate binding site in a bacterial phenylalanine hydroxylase.
Eur.Biophys.J., 42, 2013
|
|
3TK2
 
 | |
3TK4
 
 | |
