2GD4
 
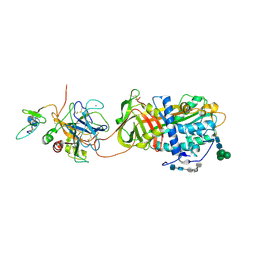 | | Crystal Structure of the Antithrombin-S195A Factor Xa-Pentasaccharide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-methyl 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranoside, ... | | Authors: | Johnson, D.J, Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2006-03-15 | | Release date: | 2006-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Antithrombin-S195A factor Xa-heparin structure reveals the allosteric mechanism of antithrombin activation.
Embo J., 25, 2006
|
|
2CFU
 
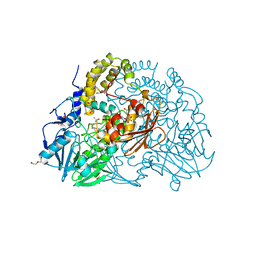 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-decane-sulfonic-acid. | | Descriptor: | 1-DECANE-SULFONIC-ACID, DI(HYDROXYETHYL)ETHER, ISOPROPYL ALCOHOL, ... | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-23 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2V3A
 
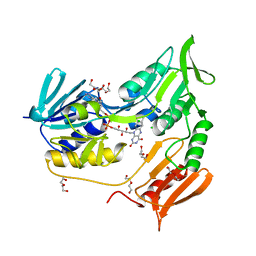 | | Crystal structure of rubredoxin reductase from Pseudomonas aeruginosa. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hagelueken, G, Wiehlmann, L, Adams, T.M, Kolmar, H, Heinz, D.W, Tuemmler, B, Schubert, W.-D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Electron Transfer Complex Rubredoxin - Rubredoxin Reductase from Pseudomonas Aeruginosa.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2V3B
 
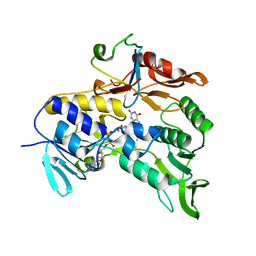 | | Crystal structure of the electron transfer complex rubredoxin - rubredoxin reductase from Pseudomonas aeruginosa. | | Descriptor: | FE (III) ION, FLAVIN-ADENINE DINUCLEOTIDE, RUBREDOXIN 2, ... | | Authors: | Hagelueken, G, Wiehlmann, L, Adams, T.M, Kolmar, H, Heinz, D.W, Tuemmler, B, Schubert, W.-D. | | Deposit date: | 2007-06-14 | | Release date: | 2007-08-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of the Electron Transfer Complex Rubredoxin - Rubredoxin Reductase from Pseudomonas Aeruginosa.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2CFZ
 
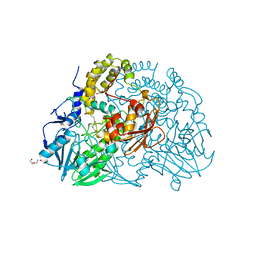 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with 1-dodecanol | | Descriptor: | 1-DODECANOL, DI(HYDROXYETHYL)ETHER, SDS HYDROLASE SDSA1, ... | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-26 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CG2
 
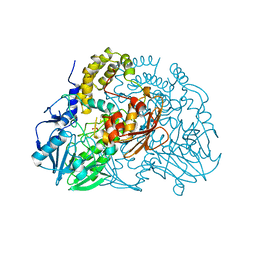 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa, in complex with sulfate | | Descriptor: | SDSA1, SULFATE ION, ZINC ION | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Class of Sulfatases.
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2CG3
 
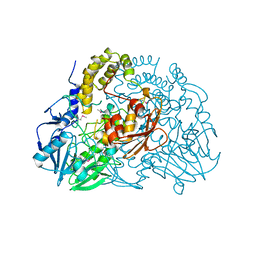 | | Crystal structure of SdsA1, an alkylsulfatase from Pseudomonas aeruginosa. | | Descriptor: | SDSA1, ZINC ION | | Authors: | Hagelueken, G, Adams, T.M, Wiehlmann, L, Widow, U, Kolmar, H, Tuemmler, B, Heinz, D.W, Schubert, W.-D. | | Deposit date: | 2006-02-27 | | Release date: | 2006-04-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Sdsa1, an Alkylsulfatase from Pseudomonas Aeruginosa, Defines a Third Third Class of Sulfatases
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
2HI9
 
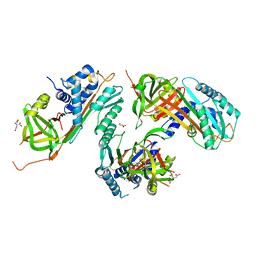 | |
6B4M
 
 | | Structural characterization of a novel monotreme-specific protein from the milk of the platypus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, IODIDE ION, ... | | Authors: | Peat, T.S, Newman, J, Sharp, J.A, Kumar, A, Nicholas, K.R, Adams, T.E. | | Deposit date: | 2017-09-27 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a novel monotreme-specific protein with antimicrobial activity from the milk of the platypus.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
6VHI
 
 | |
7K09
 
 | | Puromycin N-acetyltransferase in complex with acetyl-CoA | | Descriptor: | ACETYL COENZYME *A, Puromycin N-acetyltransferase | | Authors: | Caputo, A.T, Newman, J, Adams, T.E, Peat, T.S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
7K0A
 
 | | Puromycin N-acetyltransferase in complex with acetylated puromycin and CoA | | Descriptor: | 1,2-ETHANEDIOL, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Caputo, A.T, Newman, J, Adams, T.E. | | Deposit date: | 2020-09-03 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided selection of puromycin N-acetyltransferase mutants with enhanced selection stringency for deriving mammalian cell lines expressing recombinant proteins.
Sci Rep, 11, 2021
|
|
3KR3
 
 | | Crystal structure of IGF-II antibody complex | | Descriptor: | 1,2-ETHANEDIOL, Insulin-like growth factor II, antibody-Fab (heavy chain), ... | | Authors: | Peat, T.S, Newman, J, Adams, T.E. | | Deposit date: | 2009-11-17 | | Release date: | 2010-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A human monoclonal antibody against insulin-like growth factor-II blocks the growth of human hepatocellular carcinoma cell lines in vitro and in vivo.
Mol.Cancer Ther., 9, 2010
|
|
3B9F
 
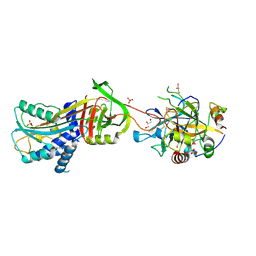 | | 1.6 A structure of the PCI-thrombin-heparin complex | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, GLYCEROL, Plasma serine protease inhibitor, ... | | Authors: | Li, W, Adams, T.E, Huntington, J.A. | | Deposit date: | 2007-11-05 | | Release date: | 2008-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-A structure of the heparin-bridged complex.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
