1P4L
 
 | | Crystal structure of NK receptor Ly49C mutant with its MHC class I ligand H-2Kb | | Descriptor: | Beta-2-microglobulin, LY49-C, MHC CLASS I H-2KB HEAVY CHAIN, ... | | Authors: | Dam, J, Guan, R, Natarajan, K, Dimasi, N, Mariuzza, R.A. | | Deposit date: | 2003-04-23 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
6PRO
 
 | | MnSOD from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Adams, J.J, Morton, C.J, Parker, M.W. | | Deposit date: | 2019-07-10 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.263 Å) | | Cite: | The Crystal Structure of the Manganese Superoxide Dismutase from Geobacillus stearothermophilus: Parker and Blake (1988) Revisited
Aust.J.Chem., 73, 2020
|
|
1MA1
 
 | | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum | | Descriptor: | FE (III) ION, superoxide dismutase | | Authors: | Adams, J.J, Anderson, B.F, Renault, J.P, Verchere-Beaur, C, Morgenstern-Badarau, I, Jameson, G.B. | | Deposit date: | 2002-07-31 | | Release date: | 2002-08-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and properties of the atypical iron superoxide dismutase from Methanobacterium thermoautotrophicum
To be published
|
|
2AEW
 
 | |
2AKQ
 
 | | The structure of bovine B-lactoglobulin A in crystals grown at very low ionic strength | | Descriptor: | Beta-lactoglobulin variant A | | Authors: | Adams, J.J, Anderson, B.F, Norris, G.E, Creamer, L.K, Jameson, G.B. | | Deposit date: | 2005-08-03 | | Release date: | 2005-08-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of bovine beta-lactoglobulin (variant A) at very low ionic strength
J.Struct.Biol., 154, 2006
|
|
2B59
 
 | |
2OZN
 
 | | The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringens | | Descriptor: | CALCIUM ION, CHLORIDE ION, Hyalurononglucosaminidase, ... | | Authors: | Adams, J.J, Boraston, A, Smith, S.P. | | Deposit date: | 2007-02-26 | | Release date: | 2008-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of Clostridium perfringens toxin complex formation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2XVS
 
 | | Crystal structure of human TTC5 (Strap) C-terminal OB domain | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, IODIDE ION, ... | | Authors: | Adams, J, Pike, A.C.W, Maniam, S, Sharpe, T.D, Coutts, A.S, Knapp, S, La Thangue, B, Bullock, A.N. | | Deposit date: | 2010-10-31 | | Release date: | 2010-11-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The P53 Cofactor Strap Exhibits an Unexpected Tpr Motif and Oligonucleotide-Binding (Ob)-Fold Structure.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6YN2
 
 | |
3KCP
 
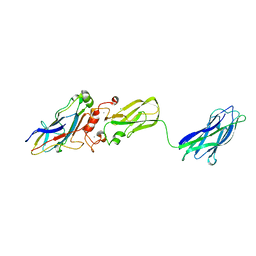 | | Crystal structure of interacting Clostridium thermocellum multimodular components | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cellulosomal-scaffolding protein A, ... | | Authors: | Adams, J.J, Currie, M.A, Bayer, E.A, Jia, Z, Smith, S.P. | | Deposit date: | 2009-10-21 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Insights into Higher-Order Organization of the Cellulosome Revealed by a Dissect-and-Build Approach: Crystal Structure of Interacting Clostridium thermocellum Multimodular Components
J.Mol.Biol., 396, 2010
|
|
2JDM
 
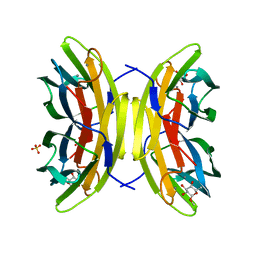 | | Mutant (S22A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDN
 
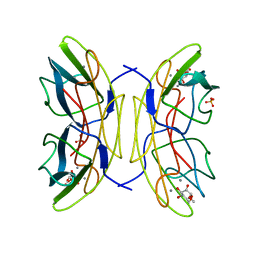 | | Mutant (S22A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-mannopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, SULFATE ION, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDU
 
 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, GLYCEROL, ... | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDY
 
 | | Mutant (G24N) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-b-D-mannoyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-D-mannopyranoside | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
2JDP
 
 | | Mutant (S23A) of Pseudomonas aeruginosa lectin II (PA-IIL) complexed with methyl-a-L-fucopyranoside | | Descriptor: | CALCIUM ION, FUCOSE-BINDING LECTIN PA-IIL, methyl alpha-L-fucopyranoside | | Authors: | Adam, J, Pokorna, M, Sabin, C, Mitchell, E.P, Imberty, A, Wimmerova, M. | | Deposit date: | 2007-01-11 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Engineering of Pa-Iil Lectin from Pseudomonas Aeruginosa - Unravelling the Role of the Specificity Loop for Sugar Preference.
Bmc Struct.Biol., 7, 2007
|
|
3TPU
 
 | | 42F3 p5E8/H2-Ld complex | | Descriptor: | 1,2-ETHANEDIOL, 42F3 alpha, 42F3 beta, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-09-08 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
2N9Q
 
 | | Photoswitchable G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*(AZW)P*GP*G)-3'), POTASSIUM ION | | Authors: | Thevarpadam, J, Bessi, I, Binas, O, Goncalves, D.P.N, Slavov, C, Jonker, H.R.A, Richter, C, Wachtveitl, J, Schwalbe, H, Heckel, A. | | Deposit date: | 2015-12-02 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Photoresponsive Formation of an Intermolecular Minimal G-Quadruplex Motif.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
3TFK
 
 | | 42F3-p4B10/H2-Ld | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.753 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TF7
 
 | | 42F3 QL9/H2-Ld complex | | Descriptor: | 42F3 Mut7 scFv (42F3 alpha chain, linker, 42F3 beta chain), ... | | Authors: | Adams, J.J, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
3TJH
 
 | | 42F3-p3A1/H2-Ld complex | | Descriptor: | 42F3 alpha, 42F3 beta, H2-Ld SBM2, ... | | Authors: | Adams, J.J, Kruse, A, Kranz, D.M, Garcia, K.C. | | Deposit date: | 2011-08-24 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | T cell receptor signaling is limited by docking geometry to peptide-major histocompatibility complex.
Immunity, 35, 2011
|
|
1P1Z
 
 | | X-RAY CRYSTAL STRUCTURE OF THE LECTIN-LIKE NATURAL KILLER CELL RECEPTOR LY-49C BOUND TO ITS MHC CLASS I LIGAND H-2Kb | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Dimasi, N, Natarajan, K, Rangjin, G, Dam, J, Margulies, D.H, Mariuzza, R.A. | | Deposit date: | 2003-04-14 | | Release date: | 2003-11-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.26 Å) | | Cite: | Variable MHC class I engagement by Ly49 natural killer cell receptors demonstrated by the crystal structure of Ly49C bound to H-2K(b).
Nat.Immunol., 4, 2003
|
|
1JLU
 
 | | Crystal Structure of the Catalytic Subunit of cAMP-dependent Protein Kinase Complexed with a Phosphorylated Substrate Peptide and Detergent | | Descriptor: | AMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR, ... | | Authors: | Madhusudan, Trafny, E.A, Xuong, N.-H, Adams, J.A, Ten Eyck, L.F, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-01 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | cAMP-dependent protein kinase: crystallographic insights into substrate recognition and phosphotransfer.
Protein Sci., 3, 1994
|
|
2V5D
 
 | | Structure of a Family 84 Glycoside Hydrolase and a Family 32 Carbohydrate-Binding Module in Tandem from Clostridium perfringens. | | Descriptor: | CALCIUM ION, O-GLCNACASE NAGJ | | Authors: | Ficko-Blean, E, Gregg, K.J, Adams, J.J, Hehemann, J.H, Smith, S.J, Czjzek, M, Boraston, A.B. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Portrait of an Enzyme: A Complete Structural Analysis of a Multi-Modular Beta-N-Acetylglucosaminidase from Clostridium Perfringens
J.Biol.Chem., 284, 2009
|
|
3ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
2IML
 
 | | Crystal structure of a hypothetical protein from Archaeoglobus fulgidus binding riboflavin 5'-phosphate | | Descriptor: | FLAVIN MONONUCLEOTIDE, GLYCEROL, Hypothetical protein | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Adams, J, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-04 | | Release date: | 2006-10-17 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a hypothetical protein from
Archaeoglobus fulgidus binding riboflavin 5'-phosphate
To be Published
|
|
