1CIM
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(AMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO (2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, METHYL MERCURY ION, ... | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
1CVE
 
 | |
3ONG
 
 | | Crystal structure of UBA2ufd-Ubc9: insights into E1-E2 interactions in Sumo pathways | | Descriptor: | SUMO-conjugating enzyme UBC9, Ubiquitin-activating enzyme E1-like | | Authors: | Wang, J, Taherbhoy, A.M, Hunt, H.W, Seyedin, S.N, Miller, D.W, Huang, D.T, Schulman, B.A. | | Deposit date: | 2010-08-28 | | Release date: | 2011-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways.
Plos One, 5, 2010
|
|
1CA3
 
 | |
3ONH
 
 | | Crystal structure of UBA2ufd-Ubc9: insights into E1-E2 interactions in Sumo pathways | | Descriptor: | Ubiquitin-activating enzyme E1-like | | Authors: | Wang, J, Taherbhoy, A.M, Hunt, H.W, Seyedin, S.N, Miller, D.W, Huang, D.T, Schulman, B.A. | | Deposit date: | 2010-08-28 | | Release date: | 2011-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of UBA2(ufd)-Ubc9: insights into E1-E2 interactions in Sumo pathways.
Plos One, 5, 2010
|
|
3P5P
 
 | | Crystal Structure of Taxadiene Synthase from Pacific Yew (Taxus brevifolia) in complex with Mg2+ and 13-aza-13,14-dihydrocopalyl diphosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-(methyl{2-[(1S,4aS,8aS)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]ethyl}amino)ethyl trihydrogen diphosphate, MAGNESIUM ION, ... | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.816 Å) | | Cite: | Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis.
Nature, 469, 2011
|
|
1CCU
 
 | |
3PE7
 
 | | Oligogalacturonate lyase in complex with manganese | | Descriptor: | ACETATE ION, CALCIUM ION, MANGANESE (II) ION, ... | | Authors: | Abbott, D.W, Gilbert, H.J, Boraston, A.B. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The active site of oligogalacturonate lyase provides unique insights into cytoplasmic oligogalacturonate beta-elimination.
J.Biol.Chem., 285, 2010
|
|
1CCT
 
 | |
1CIL
 
 | | THE POSITIONS OF HIS-64 AND A BOUND WATER IN HUMAN CARBONIC ANHYDRASE II UPON BINDING THREE STRUCTURALLY RELATED INHIBITORS | | Descriptor: | (4S-TRANS)-4-(ETHYLAMINO)-5,6-DIHYDRO-6-METHYL-4H-THIENO(2,3-B)THIOPYRAN-2-SULFONAMIDE-7,7-DIOXIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Smith, G.M, Alexander, R.S, Christianson, D.W, Mckeever, B.M, Ponticello, G.S, Springer, J.P, Randall, W.C, Baldwin, J.J, Habecker, C.N. | | Deposit date: | 1993-10-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Positions of His-64 and a bound water in human carbonic anhydrase II upon binding three structurally related inhibitors.
Protein Sci., 3, 1994
|
|
1CNG
 
 | |
1CNK
 
 | |
1CNH
 
 | |
1CPS
 
 | |
109D
 
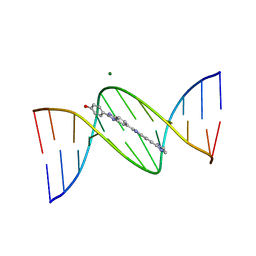 | | VARIABILITY IN DNA MINOR GROOVE WIDTH RECOGNISED BY LIGAND BINDING: THE CRYSTAL STRUCTURE OF A BIS-BENZIMIDAZOLE COMPOUND BOUND TO THE DNA DUPLEX D(CGCGAATTCGCG)2 | | Descriptor: | 5-(2-IMIDAZOLINYL)-2-[2-(4-HYDROXYPHENYL)-5-BENZIMIDAZOLYL]BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Czarny, A, Boykin, D.W, Wood, A.A, Nunn, C.M, Neidle, S, Zhao, M, Wilson, W.D. | | Deposit date: | 1995-02-15 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Variability in DNA minor groove width recognised by ligand binding: the crystal structure of a bis-benzimidazole compound bound to the DNA duplex d(CGCGAATTCGCG)2.
Nucleic Acids Res., 23, 1995
|
|
1DAN
 
 | |
3PHO
 
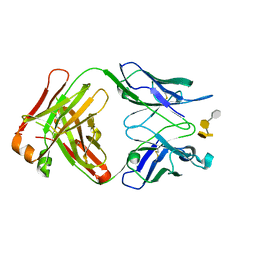 | | Crystal structure of S64-4 in complex with PSBP | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-8)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)-3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-4-O-phosphono-beta-D-glucopyranose, S64-4 Fab (IgG1) heavy chain, S64-4 Fab (IgG1) light chain | | Authors: | Evans, D.W, Evans, S.V. | | Deposit date: | 2010-11-04 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insights into parallel strategies for germline antibody recognition of lipopolysaccharide from Chlamydia.
Glycobiology, 21, 2011
|
|
12CA
 
 | |
3PND
 
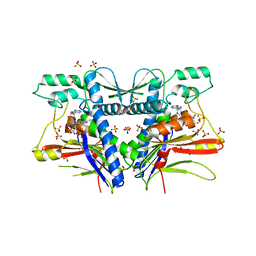 | | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD binding proteins | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Thiamine biosynthesis lipoprotein ApbE | | Authors: | Boyd, J.M, Endrizzi, J.A, Hamilton, T.L, Christopherson, M.R, Mulder, D.W, Downs, D.M, Peters, J.W. | | Deposit date: | 2010-11-18 | | Release date: | 2011-01-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | FAD binding by ApbE protein from Salmonella enterica: a new class of FAD-binding proteins.
J.Bacteriol., 193, 2011
|
|
1DI1
 
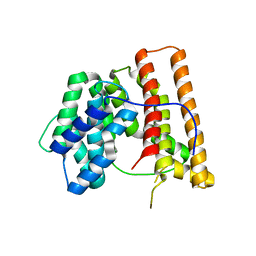 | | CRYSTAL STRUCTURE OF ARISTOLOCHENE SYNTHASE FROM PENICILLIUM ROQUEFORTI | | Descriptor: | ARISTOLOCHENE SYNTHASE | | Authors: | Caruthers, J.M, Kang, I, Cane, D.E, Christianson, D.W, Rynkiewicz, M.J. | | Deposit date: | 1999-11-28 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure determination of aristolochene synthase from the blue cheese mold, Penicillium roqueforti.
J.Biol.Chem., 275, 2000
|
|
138L
 
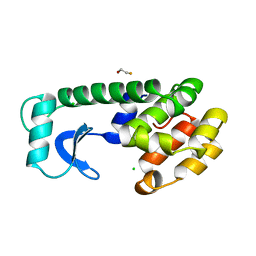 | |
139L
 
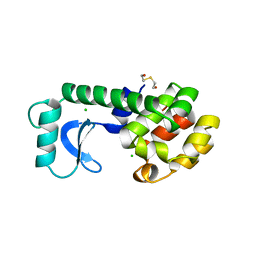 | |
1AJ8
 
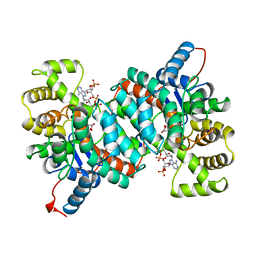 | | CITRATE SYNTHASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | CITRATE SYNTHASE, CITRIC ACID, COENZYME A | | Authors: | Russell, R.J.M, Ferguson, J.M.C, Hough, D.W, Danson, M.J, Taylor, G.L. | | Deposit date: | 1997-05-16 | | Release date: | 1997-11-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution,.
Biochemistry, 36, 1997
|
|
3PHQ
 
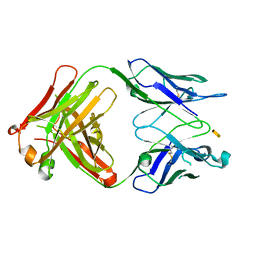 | | Crystal structure of S64-4 in complex with KDO | | Descriptor: | 3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid, S64-4 Fab (IgG1) heavy chain, S64-4 Fab (IgG1) light chain | | Authors: | Evans, D.W, Evans, S.V. | | Deposit date: | 2010-11-04 | | Release date: | 2011-05-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into parallel strategies for germline antibody recognition of lipopolysaccharide from Chlamydia.
Glycobiology, 21, 2011
|
|
1DMY
 
 | |
