1P42
 
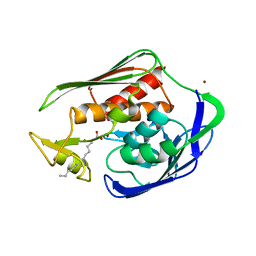 | | Crystal structure of Aquifex aeolicus LpxC Deacetylase (Zinc-Inhibited Form) | | Descriptor: | MYRISTIC ACID, UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase, ZINC ION | | Authors: | Whittington, D.A, Rusche, K.M, Shin, H, Fierke, C.A, Christianson, D.W. | | Deposit date: | 2003-04-21 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of LpxC, a Zinc-Dependent Deacetylase Essential for Endotoxin Biosynthesis
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1P8N
 
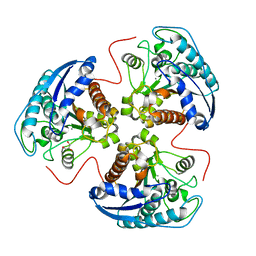 | | Structural and Functional Importance of First-Shell Metal Ligands in the Binuclear Manganese Cluster of Arginase I. | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-05-07 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional importance of first-shell metal ligands in the binuclear
manganese cluster of arginase I
Biochemistry, 42, 2003
|
|
1P8Q
 
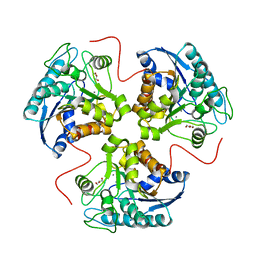 | | Structural and Functional Importance of First-Shell Metal Ligands in the Binuclear Cluster of Arginase I. | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Emig, F.A, Ash, D.E, Christianson, D.W. | | Deposit date: | 2003-05-07 | | Release date: | 2003-06-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural and functional importance of first-shell metal ligands in the binuclear
manganese cluster of arginase I
Biochemistry, 42, 2003
|
|
1T4T
 
 | | arginase-dinor-NOHA complex | | Descriptor: | 3-{[(E)-AMINO(HYDROXYIMINO)METHYL]AMINO}ALANINE, Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Shoufa, H, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1TBH
 
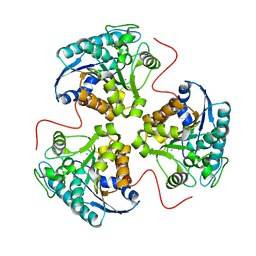 | | H141D mutant of rat liver arginase I | | Descriptor: | Arginase 1, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-20 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
1T5F
 
 | | arginase I-AOH complex | | Descriptor: | (S)-2-AMINO-7,7-DIHYDROXYHEPTANOIC ACID, Arginase 1, MANGANESE (II) ION | | Authors: | Shin, H, Cama, E, Christianson, D.W. | | Deposit date: | 2004-05-04 | | Release date: | 2005-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design of amino acid aldehydes as transition-state analogue inhibitors of arginase
J.Am.Chem.Soc., 126, 2004
|
|
1TBJ
 
 | | H141A mutant of rat liver arginase I | | Descriptor: | Arginase 1, GLYCEROL, MANGANESE (II) ION | | Authors: | Cama, E, Cox, J.D, Ash, D.E, Christianson, D.W. | | Deposit date: | 2004-05-20 | | Release date: | 2005-08-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Probing the role of the hyper-reactive histidine residue of arginase.
Arch.Biochem.Biophys., 444, 2005
|
|
1Q45
 
 | | 12-0xo-phytodienoate reductase isoform 3 | | Descriptor: | 12-oxophytodienoate-10,11-reductase, FLAVIN MONONUCLEOTIDE | | Authors: | Phillips Jr, G.N, Johnson, K.A, Bingman, C.A, Smith, D.W, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure of Arabidopsis At2g06050, 12-oxophytodienoate reductase isoform 3
Proteins, 58, 2005
|
|
1Q44
 
 | | Crystal Structure of an Arabidopsis Thaliana Putative Steroid Sulfotransferase | | Descriptor: | MALONIC ACID, Steroid Sulfotransferase | | Authors: | Phillips Jr, G.N, Smith, D.W, Johnson, K.A, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2003-08-01 | | Release date: | 2003-11-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of At2g03760, a putative steroid sulfotransferase from Arabidopsis thaliana
Proteins, 57, 2004
|
|
1PTG
 
 | | PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C IN COMPLEX WITH MYO-INOSITOL | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, PHOSPHATIDYLINOSITOL-SPECIFIC PHOSPHOLIPASE C | | Authors: | Heinz, D.W, Ryan, M, Bullock, T.L, Griffith, O.H. | | Deposit date: | 1995-05-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol.
EMBO J., 14, 1995
|
|
1PTD
 
 | |
1T4P
 
 | | Arginase-dehydro-ABH complex | | Descriptor: | Arginase 1, MANGANESE (II) ION, [(1E,5S)-5-AMINO-5-CARBOXYPENT-1-ENYL](TRIHYDROXY)BORATE(1-) | | Authors: | Cama, E, Pethe, S, Boucher, J.-L, Han, S, Emig, F.A, Ash, D.E, Viola, R.E, Mansuy, D, Christianson, D.W. | | Deposit date: | 2004-04-30 | | Release date: | 2005-04-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Inhibitor coordination interactions in the binuclear manganese cluster of arginase
Biochemistry, 43, 2004
|
|
1TS9
 
 | |
1SIB
 
 | |
1F8G
 
 | | THE X-RAY STRUCTURE OF NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE FROM RHODOSPIRILLUM RUBRUM COMPLEXED WITH NAD+ | | Descriptor: | NICOTINAMIDE NUCLEOTIDE TRANSHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Buckley, P.A, Baz Jackson, J, Schneider, T, White, S.A, Rice, D.W, Baker, P.J. | | Deposit date: | 2000-06-30 | | Release date: | 2001-06-30 | | Last modified: | 2017-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-protein recognition, hydride transfer and proton pumping in the transhydrogenase complex.
Structure Fold.Des., 8, 2000
|
|
1UGC
 
 | |
1TT5
 
 | | Structure of APPBP1-UBA3-Ubc12N26: a unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8 | | Descriptor: | Ubiquitin-conjugating enzyme E2 M, ZINC ION, amyloid protein-binding protein 1, ... | | Authors: | Huang, D.T, Miller, D.W, Mathew, R, Cassell, R, Holton, J.M, Roussel, M.F, Schulman, B.A. | | Deposit date: | 2004-06-21 | | Release date: | 2004-09-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1TSF
 
 | |
1UGD
 
 | |
1G27
 
 | | CRYSTAL STRUCTURE OF E.COLI POLYPEPTIDE DEFORMYLASE COMPLEXED WITH THE INHIBITOR BB-3497 | | Descriptor: | 2-[(FORMYL-HYDROXY-AMINO)-METHYL]-HEXANOIC ACID (1-DIMETHYLCARBAMOYL-2,2-DIMETHYL-PROPYL)-AMIDE, NICKEL (II) ION, POLYPEPTIDE DEFORMYLASE | | Authors: | Clements, J.M, Beckett, P, Brown, A, Catlin, C, Lobell, M, Palan, S, Thomas, W, Whittaker, M, Baker, P.J, Rodgers, H.F, Barynin, V, Rice, D.W, Hunter, M.G. | | Deposit date: | 2000-10-17 | | Release date: | 2001-10-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antibiotic activity and characterization of BB-3497, a novel peptide deformylase inhibitor.
Antimicrob.Agents Chemother., 45, 2001
|
|
1G52
 
 | | CARBONIC ANHYDRASE II COMPLEXED WITH 4-(AMINOSULFONYL)-N-[(2,3-DIFLUOROPHENYL)METHYL]-BENZAMIDE | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-(2,3-DIFLUORO-BENZYL)-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Kim, C.-Y, Chang, J.S, Doyon, J.B, Baird Jr, T.T, Fierke, C.A, Jain, A, Christianson, D.W. | | Deposit date: | 2000-10-30 | | Release date: | 2000-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of Fluorine to Protein-Ligand Affinity in the Binding of Fluoroaromatic Inhibitors to Carbonic Anhydrase II
J.Am.Chem.Soc., 122, 2000
|
|
1YXE
 
 | | Structure and inter-domain interactions of domain II from the blood stage malarial protein, apical membrane antigen 1 | | Descriptor: | apical membrane antigen 1 | | Authors: | Feng, Z.P, Keizer, D.W, Stevenson, R.A, Yao, S, Babon, J.J, Murphy, V.J, Anders, R.F, Norton, R.S. | | Deposit date: | 2005-02-21 | | Release date: | 2005-07-05 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure and Inter-domain Interactions of Domain II from the Blood-stage Malarial Protein, Apical Membrane Antigen 1.
J.Mol.Biol., 350, 2005
|
|
1YYQ
 
 | | Y305F Trichodiene Synthase complexed with pyrophosphate | | Descriptor: | MAGNESIUM ION, PYROPHOSPHATE 2-, Trichodiene synthase | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
1HMV
 
 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
5THT
 
 | | Crystal Structure of G303A HDAC8 in complex with M344 | | Descriptor: | 1,2-ETHANEDIOL, 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2016-09-30 | | Release date: | 2016-12-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.407 Å) | | Cite: | Structural and Functional Influence of the Glycine-Rich Loop G302GGGY on the Catalytic Tyrosine of Histone Deacetylase 8.
Biochemistry, 55, 2016
|
|
