8F4C
 
 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4I
 
 | | RT XFEL structure of Photosystem II 2000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4K
 
 | | RT XFEL structure of the three-flash state of Photosystem II (3F, S0-rich) at 2.16 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
6P9A
 
 | | HIV-1 Protease multiple mutant PRS5B with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease, PHOSPHATE ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
Febs J., 287, 2020
|
|
4GHL
 
 | | Structural Basis for Marburg virus VP35 mediate immune evasion mechanisms | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Ramanan, P, Borek, D.M, Otwinowski, Z, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2012-08-07 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for Marburg virus VP35-mediated immune evasion mechanisms.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
8F4D
 
 | | RT XFEL structure of Photosystem II 50 microseconds after the third illumination at 2.15 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4E
 
 | | RT XFEL structure of Photosystem II 250 microseconds after the third illumination at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8F4G
 
 | | RT XFEL structure of Photosystem II 730 microseconds after the third illumination at 2.03 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
3G1R
 
 | | Crystal structure of human liver 5beta-reductase (AKR1D1) in complex with NADP and Finasteride. Resolution 1.70 A | | Descriptor: | (4aR,4bS,6aS,7S,9aS,9bS,11aR)-N-tert-butyl-4a,6a-dimethyl-2-oxo-2,4a,4b,5,6,6a,7,8,9,9a,9b,10,11,11a-tetradecahydro-1H-indeno[5,4-f]quinoline-7-carboxamide, 3-oxo-5-beta-steroid 4-dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Di Costanzo, L, Drury, J.E, Penning, T.M, Christianson, D.W. | | Deposit date: | 2009-01-30 | | Release date: | 2009-06-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Inhibition of human steroid 5beta-reductase (AKR1D1) by finasteride and structure of the enzyme-inhibitor complex.
J.Biol.Chem., 284, 2009
|
|
8F4J
 
 | | RT XFEL structure of Photosystem II 4000 microseconds after the third illumination at 2.00 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Bhowmick, A, Hussein, R, Bogacz, I, Simon, P.S, Ibrahim, M, Chatterjee, R, Doyle, M.D, Cheah, M.H, Fransson, T, Chernev, P, Kim, I.-S, Makita, H, Dasgupta, M, Kaminsky, C.J, Zhang, M, Gatcke, J, Haupt, S, Nangca, I.I, Keable, S.M, Aydin, O, Tono, K, Owada, S, Gee, L.B, Fuller, F.D, Batyuk, A, Alonso-Mori, R, Holton, J.M, Paley, D.W, Moriarty, N.W, Mamedov, F, Adams, P.D, Brewster, A.S, Dobbek, H, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2022-11-10 | | Release date: | 2023-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural evidence for intermediates during O 2 formation in photosystem II.
Nature, 617, 2023
|
|
8EQI
 
 | |
3G4F
 
 | | Crystal Structure of (+)- -Cadinene Synthase from Gossypium arboreum in complex with 2-fluorofarnesyl diphosphate | | Descriptor: | (+)-delta-cadinene synthase isozyme XC1, (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, BETA-MERCAPTOETHANOL, ... | | Authors: | Gennadios, H.A, Di Costanzo, L, Christianson, D.W. | | Deposit date: | 2009-02-03 | | Release date: | 2009-06-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis.
Biochemistry, 48, 2009
|
|
6PIC
 
 | |
6M5H
 
 | |
6M5P
 
 | | A class C beta-lactamase | | Descriptor: | (4R,5S)-3-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-5-[(2S,3R)-3-hydroxy-1-oxobutan-2-yl]-4-methyl-4,5-d ihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Bae, D.W, Jung, Y.E, Cha, S.S. | | Deposit date: | 2020-03-11 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Novel inhibition mechanism of carbapenems on the ACC-1 class C beta-lactamase.
Arch.Biochem.Biophys., 693, 2020
|
|
6MDR
 
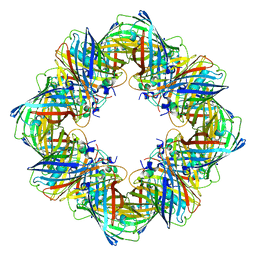 | | Cryo-EM structure of the Ceru+32/GFP-17 protomer | | Descriptor: | Ceru+32, GFP-17 | | Authors: | Simon, A.J, Zhou, Y, Ramasubramani, V, Glaser, J, Pothukuchy, A, Golihar, J, Gerberich, J.C, Leggere, J.C, Morrow, B.R, Jung, C, Glotzer, S.C, Taylor, D.W, Ellington, A.D. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-23 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Supercharging enables organized assembly of synthetic biomolecules.
Nat Chem, 11, 2019
|
|
3FI7
 
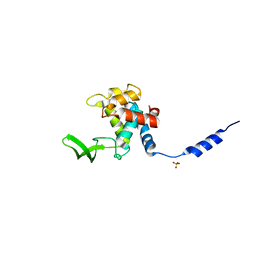 | | Crystal Structure of the autolysin Auto (Lmo1076) from Listeria monocytogenes, catalytic domain | | Descriptor: | Lmo1076 protein, SULFATE ION | | Authors: | Bublitz, M, Polle, L, Holland, C, Nimtz, M, Heinz, D.W, Schubert, W.D. | | Deposit date: | 2008-12-11 | | Release date: | 2009-04-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for autoinhibition and activation of Auto, a virulence-associated peptidoglycan hydrolase of Listeria monocytogenes.
Mol.Microbiol., 71, 2009
|
|
6PZS
 
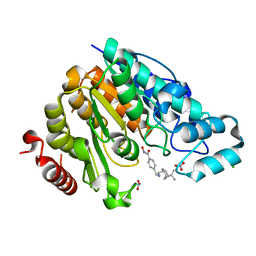 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with JR005 | | Descriptor: | 4-[({[(1R,2R,5R)-6,6-dimethylbicyclo[3.1.1]heptan-2-yl]methyl}amino)methyl]-N-hydroxybenzamide, CHLORIDE ION, Hdac6 protein, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-08-01 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Exploring Structural Determinants of Inhibitor Affinity and Selectivity in Complexes with Histone Deacetylase 6.
J.Med.Chem., 63, 2020
|
|
6PYE
 
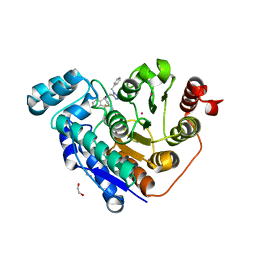 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 complexed with NR160 | | Descriptor: | 1,2-ETHANEDIOL, Hdac6 protein, N-[(1-benzyl-1H-tetrazol-5-yl)methyl]-N-{[4-(hydroxycarbamoyl)phenyl]methyl}-2-(trifluoromethyl)benzamide, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-07-29 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.480003 Å) | | Cite: | Multicomponent Synthesis, Binding Mode, and Structure-Activity Relationship of Selective Histone Deacetylase 6 (HDAC6) Inhibitors with Bifurcated Capping Groups.
J.Med.Chem., 63, 2020
|
|
6PZR
 
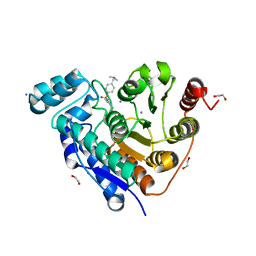 | |
6Q0Z
 
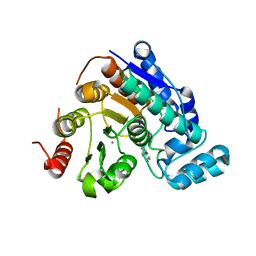 | |
3FKE
 
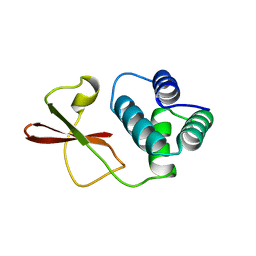 | | Structure of the Ebola VP35 Interferon Inhibitory Domain | | Descriptor: | Polymerase cofactor VP35 | | Authors: | Amarasinghe, G.K, Leung, D.W, Ginder, N.D, Honzatko, R.B, Nix, J, Basler, C.F, Fulton, D.B. | | Deposit date: | 2008-12-16 | | Release date: | 2009-01-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the Ebola VP35 interferon inhibitory domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6LTQ
 
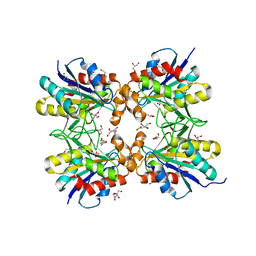 | | Crystal structure of pyrrolidone carboxyl peptidase from thermophilic keratin degrading bacterium Fervidobacterium islandicum AW-1 (FiPcp) | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Pyroglutamyl-peptidase I | | Authors: | Dhanasingh, I, Jin, H.S, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-01-23 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of oxidized pyrrolidone carboxypeptidase from Fervidobacterium islandicum AW-1 reveals unique structural features for thermostability and keratinolysis.
Biochem.Biophys.Res.Commun., 540, 2021
|
|
3FIB
 
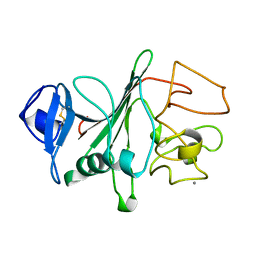 | | RECOMBINANT HUMAN GAMMA-FIBRINOGEN CARBOXYL TERMINAL FRAGMENT (RESIDUES 143-411) BOUND TO CALCIUM AT PH 6.0: A FURTHER REFINEMENT OF PDB ENTRY 1FIB, AND DIFFERS FROM 1FIB BY THE MODELLING OF A CIS PEPTIDE BOND BETWEEN RESIDUES K338 AND C339 | | Descriptor: | CALCIUM ION, FIBRINOGEN GAMMA CHAIN RESIDUES | | Authors: | Pratt, K.P, Cote, H.C.F, Chung, D.W, Stenkamp, R.E, Davie, E.W. | | Deposit date: | 1997-07-14 | | Release date: | 1997-09-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
6M5C
 
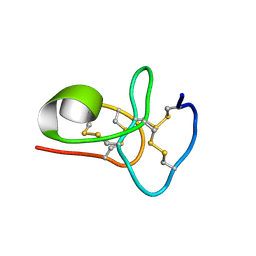 | | Solution structure of avenatide aV1 | | Descriptor: | avenatide aV1 | | Authors: | Tay, S.V, Wong, K.H, Huang, J.Y, Fan, J.S, Yang, D.W, Tam, J.P. | | Deposit date: | 2020-03-10 | | Release date: | 2021-03-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of avenatide aV1
To Be Published
|
|
