3J8F
 
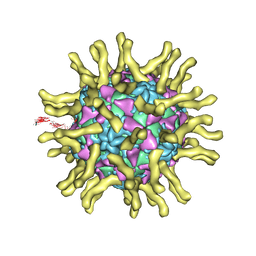 | | Cryo-EM reconstruction of poliovirus-receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3GU5
 
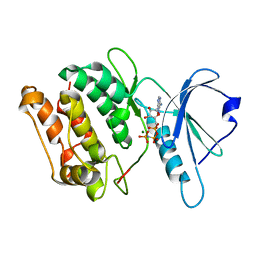 | |
3HJU
 
 | | Crystal structure of human monoglyceride lipase | | Descriptor: | GLYCEROL, Monoglyceride lipase | | Authors: | Labar, G, Bauvois, C, Borel, F, Ferrer, J.-L, Wouters, J, Lambert, D.M. | | Deposit date: | 2009-05-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Human Monoacylglycerol Lipase, a Key Actor in Endocannabinoid Signaling
Chembiochem, 11, 2010
|
|
3J3O
 
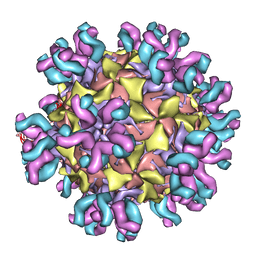 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 160S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
3I5H
 
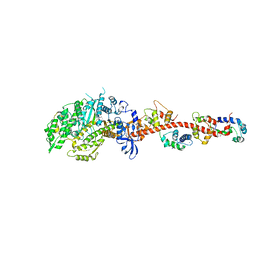 | | The crystal structure of rigor like squid myosin S1 in the absence of nucleotide | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3HS5
 
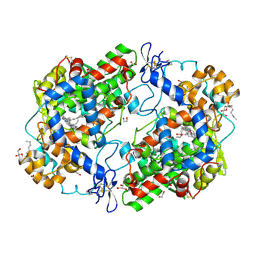 | | X-ray crystal structure of arachidonic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3HS6
 
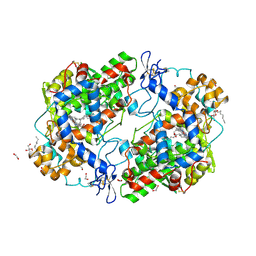 | | X-ray crystal structure of eicosapentaenoic acid bound to the cyclooxygenase channel of cyclooxygenase-2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-06-10 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
3HSL
 
 | | The Crystal Structure of PF-8, the DNA Polymerase Accessory Subunit from Kaposi's Sarcoma-Associated Herpesvirus | | Descriptor: | ORF59 | | Authors: | Baltz, J.L, Filman, D.J, Ciustea, M, Silverman, J.E.Y, Lautenschlager, C.L, Coen, D.M, Ricciardi, R.P, Hogle, J.M. | | Deposit date: | 2009-06-10 | | Release date: | 2009-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of PF-8, the DNA polymerase accessory subunit from Kaposi's sarcoma-associated herpesvirus.
J.Virol., 83, 2009
|
|
3JTD
 
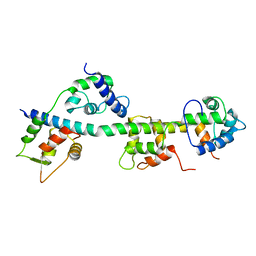 | | Calcium-free Scallop Myosin Regulatory Domain with ELC-D19A Point Mutation | | Descriptor: | MAGNESIUM ION, Myosin essential light chain, striated adductor muscle, ... | | Authors: | Himmel, D.M, Mui, S, O'Neall-Hennessey, E, Szent-Gyorgyi, A, Cohen, C. | | Deposit date: | 2009-09-11 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | The on-off switch in regulated myosins: different triggers but related mechanisms.
J.Mol.Biol., 394, 2009
|
|
3K2P
 
 | | HIV-1 Reverse Transcriptase Isolated RnaseH Domain with the Inhibitor beta-thujaplicinol Bound at the Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, Reverse Transcriptase | | Authors: | Pauly, T.A, Himmel, D.M, Maegley, K, Arnold, E. | | Deposit date: | 2009-09-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
5TCV
 
 | |
5TCW
 
 | |
5THE
 
 | |
5UA8
 
 | | Ocellatin-F1, solution structure in SDS micelle by NMR spectroscopy | | Descriptor: | Ocellatin-F1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5UA6
 
 | | Ocellatin-LB1, solution structure in SDS micelle by NMR spectroscopy | | Descriptor: | Ocellatin-LB1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5U9R
 
 | | Ocellatin-LB2, solution structure in TFE by NMR spectroscopy | | Descriptor: | Ocellatin-LB2 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, Verly, R.M, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-29 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5U9V
 
 | | Ocellatin-LB1, solution structure in DPC micelle by NMR spectroscopy | | Descriptor: | Ocellatin-LB1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-29 | | Method: | SOLUTION NMR | | Cite: | Ocellatin-LB1
To Be Published
|
|
5UA7
 
 | | Ocellatin-LB2, solution structure in SDS micelle by NMR spectroscopy | | Descriptor: | Ocellatin-LB2 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-19 | | Release date: | 2017-03-29 | | Last modified: | 2018-04-18 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5UEN
 
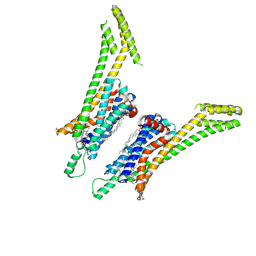 | | Crystal structure of the human adenosine A1 receptor A1AR-bRIL in complex with the covalent antagonist DU172 at 3.2A resolution | | Descriptor: | 4-{[3-(8-cyclohexyl-2,6-dioxo-1-propyl-1,2,6,7-tetrahydro-3H-purin-3-yl)propyl]carbamoyl}benzene-1-sulfonyl fluoride, Adenosine receptor A1,Soluble cytochrome b562,Adenosine receptor A1, OLEIC ACID | | Authors: | Glukhova, A, Thal, D.M, Nguyen, A.T, Vecchio, E.A, Jorg, M, Scammells, P.J, May, L.T, Sexton, P.M, Christopoulos, A. | | Deposit date: | 2017-01-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of the Adenosine A1 Receptor Reveals the Basis for Subtype Selectivity.
Cell, 168, 2017
|
|
5U9S
 
 | | Ocellatin-F1, solution structure in TFE by NMR spectroscopy | | Descriptor: | Ocellatin-F1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ocellatin-F1
To Be Published
|
|
5U9Y
 
 | | Ocellatin-F1 | | Descriptor: | Ocellatin-K1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | NMR structures in different membrane environments of three ocellatin peptides isolated from Leptodactylus labyrinthicus.
Peptides, 103, 2018
|
|
5U9X
 
 | | Ocellatin-LB2 | | Descriptor: | Ocellatin-K1 | | Authors: | Gusmao, K.A.G, dos Santos, D.M, Santos, V.M, Pilo-Veloso, D, de Lima, M.E, Resende, J.M. | | Deposit date: | 2016-12-18 | | Release date: | 2017-03-01 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Ocellatin-LB2
To Be Published
|
|
5USH
 
 | | Structure of vaccinia virus D8 protein bound to human Fab vv66 | | Descriptor: | Fab vv66 heavy chain, Fab vv66 light chain, IMV membrane protein | | Authors: | Zajonc, D.M. | | Deposit date: | 2017-02-13 | | Release date: | 2017-09-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-function characterization of three human antibodies targeting the vaccinia virus adhesion molecule D8.
J. Biol. Chem., 293, 2018
|
|
5UZ7
 
 | | Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex | | Descriptor: | Calcitonin receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liang, Y.L, Khoshouei, M, Radjainia, M, Zhang, Y, Glukhova, A, Tarrasch, J, Thal, D.M, Furness, S.G.B, Christopoulos, G, Coudrat, T, Danev, R, Baumeister, W, Miller, L.J, Christopoulos, A, Kobilka, B.K, Wootten, D, Skiniotis, G, Sexton, P.M. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Phase-plate cryo-EM structure of a class B GPCR-G-protein complex.
Nature, 546, 2017
|
|
5V8S
 
 | | Flavo di-iron protein H90D mutant from Thermotoga maritima | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Taylor, A.B, Becker, A, Giri, N, Hart, P.J, Kurtz Jr, D.M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Flavo Di-iron protein H90D Mutant from Thermotoga Maritima
To Be Published
|
|
