7OYL
 
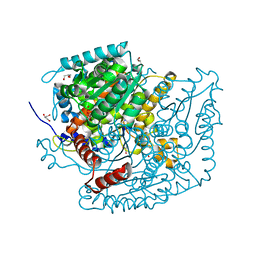 | | Phosphoglucose isomerase of Aspergillus fumigatus in complexed with Glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Raimi, O.G, Yan, K, Fang, W, van Aalten, D.M.F. | | Deposit date: | 2021-06-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Phosphoglucose Isomerase Is Important for Aspergillus fumigatus Cell Wall Biogenesis.
Mbio, 13, 2022
|
|
7P5O
 
 | |
6YL5
 
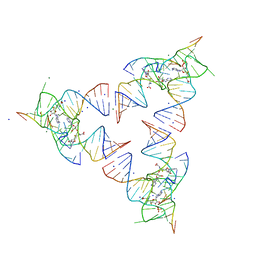 | | Crystal structure of the SAM-SAH riboswitch with SAH | | Descriptor: | Chains: A,B,C,D,E,F,G,H,I,J,K,L, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YMI
 
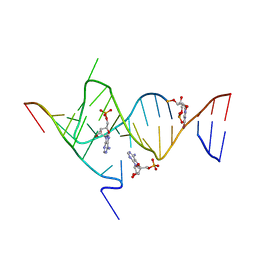 | | Crystal structure of the SAM-SAH riboswitch with AMP. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE MONOPHOSPHATE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMJ
 
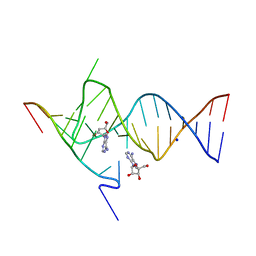 | | Crystal structure of the SAM-SAH riboswitch with adenosine. | | Descriptor: | 5-BROMOCYTIDINE 5'-(DIHYDROGEN PHOSPHATE), ADENOSINE, Chains: A,C,F,I,M,O, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
7PIZ
 
 | |
6YLB
 
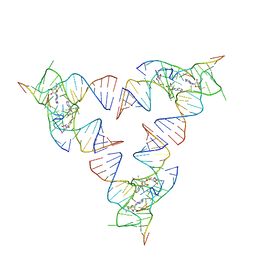 | | Crystal structure of the SAM-SAH riboswitch with SAM | | Descriptor: | Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, S-ADENOSYLMETHIONINE | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 2020
|
|
6YML
 
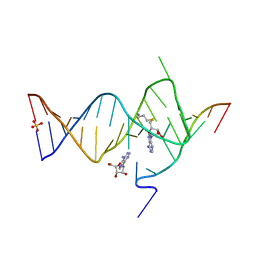 | |
7PJC
 
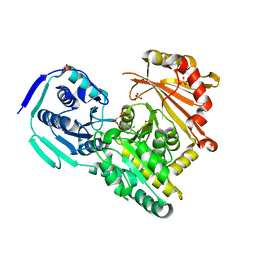 | |
7PPR
 
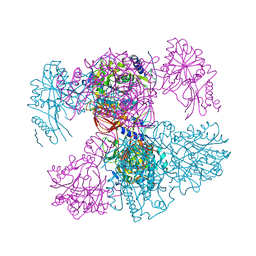 | |
6YMK
 
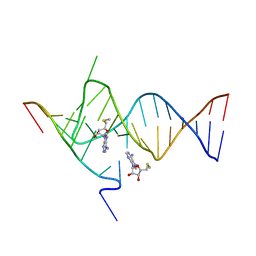 | | Crystal structure of the SAM-SAH riboswitch with AMP | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, Chains: A,C,F,I,M,O, Chains: B,D,G,J,N,P, ... | | Authors: | Huang, L, Lilley, D.M.J. | | Deposit date: | 2020-04-08 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structure and ligand-induced folding of the SAM/SAH riboswitch.
Nucleic Acids Res., 48, 2020
|
|
6YMM
 
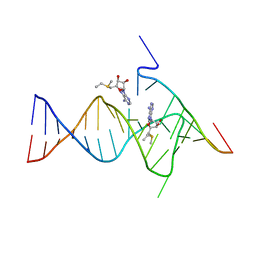 | |
4OJI
 
 | | Crystal Structure of Twister Ribozyme | | Descriptor: | MAGNESIUM ION, RNA (52-MER) | | Authors: | Liu, Y, Wilson, T.J, McPhee, S.A, Lilley, D.M.J. | | Deposit date: | 2014-01-21 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and mechanistic investigation of the twister ribozyme.
Nat.Chem.Biol., 10, 2014
|
|
6BZ5
 
 | |
6VWB
 
 | | Solution structure of the N-terminal helix-hairpin-helix domain of human MUS81 | | Descriptor: | Crossover junction endonuclease MUS81 | | Authors: | Payliss, B, Houliston, S, Lemak, A, Arrowsmith, C.H, Wyatt, H.D.M. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Phosphorylation of the DNA repair scaffold SLX4 drives folding of the SAP domain and activation of the MUS81-EME1 endonuclease
Cell Rep, 41, 2022
|
|
5LRV
 
 | | Structure of Cezanne/OTUD7B OTU domain bound to Lys11-linked diubiquitin | | Descriptor: | GLYCEROL, OTU domain-containing protein 7B, PHOSPHATE ION, ... | | Authors: | Mevissen, T.E.T, Kulathu, Y, Mulder, M.P.C, Geurink, P.P, Maslen, S.L, Gersch, M, Elliott, P.R, Burke, J.E, van Tol, B.D.M, Akutsu, M, El Oualid, F, Kawasaki, M, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2016-08-22 | | Release date: | 2016-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne.
Nature, 538, 2016
|
|
5LQT
 
 | |
4EZA
 
 | | Crystal structure of the atypical phosphoinositide (aPI) binding domain of IQGAP2 | | Descriptor: | Ras GTPase-activating-like protein IQGAP2 | | Authors: | Van Aalten, D.M.F, Dixon, M.J, Gray, A, Schenning, M, Agacan, M, Leslie, N.R, Downes, C.P, Batty, I.H, Nedyalkova, L, Tempel, W, Tong, Y, Zhong, N, Crombet, L, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | IQGAP Proteins Reveal an Atypical Phosphoinositide (aPI) Binding Domain with a Pseudo C2 Domain Fold.
J.Biol.Chem., 287, 2012
|
|
1H0G
 
 | | Complex of a chitinase with the natural product cyclopentapeptide argadin from Clonostachys | | Descriptor: | Argadin, CHITINASE B, GLYCEROL | | Authors: | Houston, D, Shiomi, K, Arai, N, Omura, S, Peter, M.G, Turberg, A, Synstad, B, Eijsink, V.G.H, Aalten, D.M.F. | | Deposit date: | 2002-06-19 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution Inhibited Complexes of a Chitinase with Natural Product Cyclopentapeptides - Peptide Mimicry of a Carbohydrate Substrate
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1GUV
 
 | | Structure of human chitotriosidase | | Descriptor: | 1,2-ETHANEDIOL, CHITOTRIOSIDASE | | Authors: | Von Moeller, H, Houston, D, Boot, R.G, Aerts, J.M.F.G, Van Aalten, D.M.F. | | Deposit date: | 2002-01-31 | | Release date: | 2003-03-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of Human Chitotriosidase - Implications for Specific Inhibitor Design and Function of Mammalian Chitinase-Like Lectins
J.Biol.Chem., 277, 2002
|
|
6IAR
 
 | | Tricyclic indazoles a novel class of selective estrogen receptor degrader antagonists | | Descriptor: | 3-[4-[(6~{R})-7-(2-methylpropyl)-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]phenyl]propanoic acid, Estrogen receptor | | Authors: | Scott, J.S, Bailey, A, Buttar, D, Carbajo, R.J, Curwen, J, Davies, R.D.M, Degorce, S.L, Donald, C, Gangl, E, Greenwood, R, Groombridge, S.D, Johnson, T, Lamont, S, Lawson, M, Lister, A, Morrow, C, Moss, T, Pink, J.H, Polanski, R. | | Deposit date: | 2018-11-27 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Tricyclic Indazoles-A Novel Class of Selective Estrogen Receptor Degrader Antagonists.
J.Med.Chem., 62, 2019
|
|
1GSW
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GSX
 
 | | CRYSTAL STRUCTURE OF THE P65 CRYSTAL FORM OF PHOTOACTIVE YELLOW PROTEIN G47S/G51S MUTANT | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Van Aalten, D.M.F, Crielaard, W, Hellingwerf, K.J, Joshua-Tor, L. | | Deposit date: | 2002-01-09 | | Release date: | 2002-02-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Engineering Photocycle Dynamics: Crystal Structures and Kinetics of Three Photoactive Yellow Protein Hinge-Bending Mutants
J.Biol.Chem., 227, 2002
|
|
1GOI
 
 | | Crystal structure of the D140N mutant of chitinase B from Serratia marcescens at 1.45 A resolution | | Descriptor: | CHITINASE B, GLYCEROL, SULFATE ION | | Authors: | Kolstad, G, Synstad, B, Eijsink, V.G.H, Van Aalten, D.M.F. | | Deposit date: | 2001-10-21 | | Release date: | 2001-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the D140N Mutant of Chitinase B from Serratia Marcescens at 1.45 A Resolution.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1H9G
 
 | | FadR, FATTY ACID RESPONSIVE TRANSCRIPTION FACTOR FROM E. COLI, in complex with myristoyl-CoA | | Descriptor: | COENZYME A, FATTY ACID METABOLISM REGULATOR PROTEIN, MYRISTIC ACID | | Authors: | Van Aalten, D.M.F, Dirusso, C.C, Knudsen, J. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis of Acyl Coenzyme A-Dependent Regulation of the Transcription Factor Fadr
Embo J., 20, 2001
|
|
