6YD9
 
 | | Ecoli GyrB24 with inhibitor 16a | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, N-[6-(3-azanylpropanoylamino)-1,3-benzothiazol-2-yl]-3,4-bis(chloranyl)-5-methyl-1H-pyrrole-2-carboxamide | | Authors: | Barancokova, M, Skok, Z, Benek, O, Cruz, C.D, Tammela, P, Tomasic, T, Zidar, N, Masic, L.P, Zega, A, Stevenson, C.E.M, Mundy, J, Lawson, D.M, Maxwell, A.M, Kikelj, D, Ilas, J. | | Deposit date: | 2020-03-20 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploring the Chemical Space of Benzothiazole-Based DNA Gyrase B Inhibitors.
Acs Med.Chem.Lett., 11, 2020
|
|
6SRZ
 
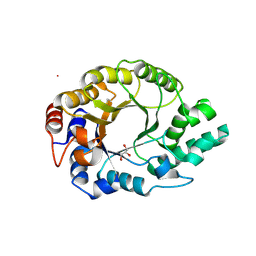 | | Kemp Eliminase HG3.17 mutant Q50H, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50H, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6SS1
 
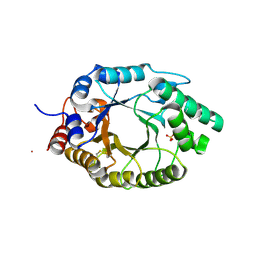 | | Kemp Eliminase HG3.17 mutant Q50A, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50A, E47N,N300D Complexed with Transition State Analog 6-Nitrobenzotriazole, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6T1F
 
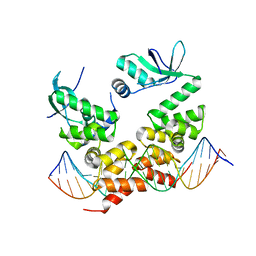 | | Crystal structure of the C-terminally truncated chromosome-partitioning protein ParB from Caulobacter crescentus complexed to the centromeric parS site | | Descriptor: | Chromosome-partitioning protein ParB, DNA (5'-D(*GP*GP*AP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*TP*CP*C)-3') | | Authors: | Jalal, A.S.B, Pastrana, C.L, Tran, N.T, Stevenson, C.E.M, Lawson, D.M, Moreno-Herrero, F, Le, T.B.K. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A CTP-dependent gating mechanism enables ParB spreading on DNA.
Elife, 10, 2021
|
|
6SS3
 
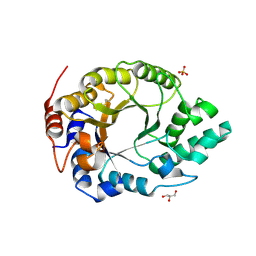 | | Kemp Eliminase HG3.17 mutant Q50K, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, GLYCEROL, Kemp Eliminase HG3.17 Q50K, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6IDY
 
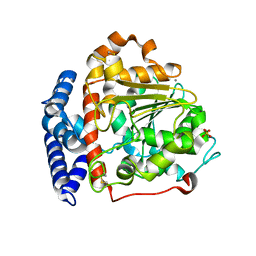 | |
6R76
 
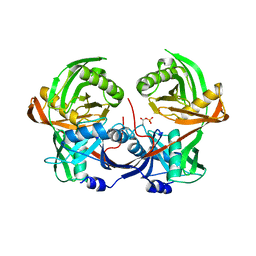 | |
5Z49
 
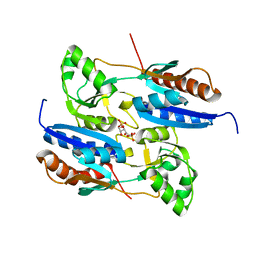 | | Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose-1,5-bisphosphate | | Descriptor: | HTH-type transcriptional activator CmpR, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Jiang, Y.L, Mahounga, D.M, Sun, H. | | Deposit date: | 2018-01-10 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Crystal structure of the effector-binding domain of Synechococcus elongatus CmpR in complex with ribulose 1,5-bisphosphate.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5X6V
 
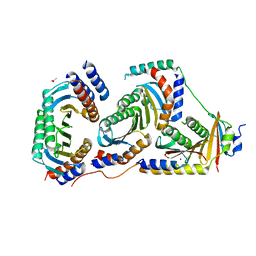 | | Crystal structure of human heteroheptameric complex | | Descriptor: | ACETATE ION, Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, ... | | Authors: | Yonehara, R, Nada, S, Nakai, T, Nakai, M, Kitamura, A, Ogawa, A, Nakatsumi, H, Nakayama, K.I, Li, S, Standley, D.M, Yamashita, E, Nakagawa, A, Okada, M. | | Deposit date: | 2017-02-23 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the assembly of the Ragulator-Rag GTPase complex.
Nat Commun, 8, 2017
|
|
6Y93
 
 | | Crystal structure of the DNA-binding domain of the Nucleoid Occlusion Factor (Noc) complexed to the Noc-binding site (NBS) | | Descriptor: | Noc Binding Site (NBS), Nucleoid occlusion protein | | Authors: | Jalal, A.S.B, Tran, N.T, Stevenson, C.E.M, Chan, E, Lo, R, Tan, X, Noy, A, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2020-03-06 | | Release date: | 2020-08-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Diversification of DNA-Binding Specificity by Permissive and Specificity-Switching Mutations in the ParB/Noc Protein Family.
Cell Rep, 32, 2020
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6SRY
 
 | | Kemp Eliminase HG3.17 mutant Q50S, E47N, N300D Complexed with Transition State Analog 6-Nitrobenzotriazole | | Descriptor: | 6-NITROBENZOTRIAZOLE, Kemp Eliminase HG3.17 Q50S, E47N,N300D, ... | | Authors: | Bloch, J.S, Pinkas, D.M, Hilvert, D. | | Deposit date: | 2019-09-06 | | Release date: | 2020-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Contribution of Oxyanion Stabilization to Kemp Eliminase Efficiencyproficiency
Acs Catalysis, 2020
|
|
6HQ8
 
 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 with laminarihexaose bound at a surface site | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
6S2Y
 
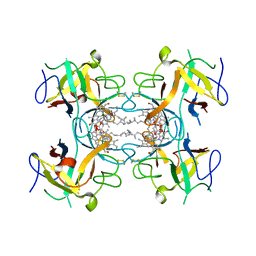 | | Water-soluble Chlorophyll Protein (WSCP) from Lepidium virginicum with Chlorophyll-b | | Descriptor: | CHLOROPHYLL B, Water-soluble chlorophyll protein | | Authors: | Agostini, A, Meneghin, E, Gewehr, L, Pedron, D, Palm, D.M, Carbonera, D, Paulsen, H, Jaenicke, E, Collini, E. | | Deposit date: | 2019-06-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How water-mediated hydrogen bonds affect chlorophyll a/b selectivity in Water-Soluble Chlorophyll Protein.
Sci Rep, 9, 2019
|
|
6HP9
 
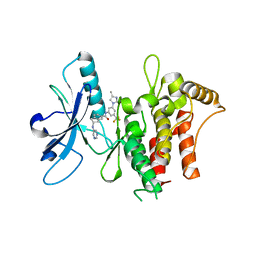 | | Structure of the kinase domain of human DDR1 in complex with a 2-Amino-2,3-Dihydro-1H-Indene-5-Carboxamide-based inhibitor | | Descriptor: | (2~{R})-~{N}-[3-(4-methylimidazol-1-yl)-5-(trifluoromethyl)phenyl]-2-(pyrimidin-5-ylamino)-2,3-dihydro-1~{H}-indene-5-carboxamide, Epithelial discoidin domain-containing receptor 1 | | Authors: | Pinkas, D.M, Fox, A.E, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-09-19 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | 2-Amino-2,3-dihydro-1H-indene-5-carboxamide-Based Discoidin Domain Receptor 1 (DDR1) Inhibitors: Design, Synthesis, and in Vivo Antipancreatic Cancer Efficacy.
J.Med.Chem., 62, 2019
|
|
6S1F
 
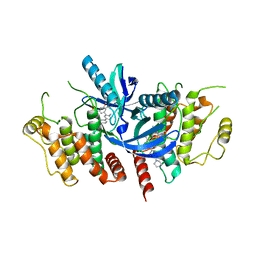 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[3-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-5-methoxy-phenyl]methanesulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of 3,5-diaryl-2-aminopyridines as receptor-interacting protein kinase 2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling inhibitors
To be published
|
|
6HQ6
 
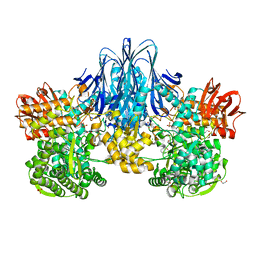 | | Bacterial beta-1,3-oligosaccharide phosphorylase from GH149 | | Descriptor: | 1,2-ETHANEDIOL, BICINE, Bacterial beta-1,3-oligosaccharide phosphorylase, ... | | Authors: | Kuhaudomlarp, S, Stevenson, C.E.M, Lawson, D.M, Field, R.A. | | Deposit date: | 2018-09-24 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of a GH149 beta-(1 → 3) glucan phosphorylase reveals a new surface oligosaccharide binding site and additional domains that are absent in the disaccharide-specific GH94 glucose-beta-(1 → 3)-glucose (laminaribiose) phosphorylase.
Proteins, 87, 2019
|
|
6T3E
 
 | | Structure of Thermococcus litoralis Delta(1)-pyrroline-2-carboxylate reductase in complex with NADH and L-proline | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DELTA1-pyrroline-2-carboxylate reductase, PROLINE | | Authors: | Ferraris, D.M, Miggiano, R, Ferrario, E, Rizzi, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of Thermococcus litoralis Delta1-pyrroline-2-carboxylate reductase in complex with NADH and L-proline.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6HQ9
 
 | | Crystal structure of the Tudor domain of human ERCC6-L2 | | Descriptor: | DNA excision repair protein ERCC-6-like 2 | | Authors: | Newman, J.A, Gavard, A.E, Nathan, W.J, Pinkas, D.M, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2018-09-24 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Crystal structure of the Tudor domain of human ERCC6-L2
To Be Published
|
|
5X61
 
 | | Crystal structure of Acetylcholinesterase Catalytic Subunit of the Malaria Vector Anopheles Gambiae, 3.4 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AMMONIUM ION, ... | | Authors: | Han, Q, Robinson, H, Ding, H, Wong, D.M, Lam, P.C.H, Totrov, M.M, Carlier, P.R, Li, J. | | Deposit date: | 2017-02-20 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of acetylcholinesterase catalytic subunits of the malaria vector Anopheles gambiae
Insect Sci., 2017
|
|
5XKS
 
 | |
6Z1A
 
 | | Ternary complex of Staphylococcus aureus DNA gyrase with AMK12 and DNA | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kolaric, A, Germe, T, Hrast, M, Stevenson, C.E.M, Lawson, D.M, Burton, N, Voros, J, Maxwell, A, Minovski, N, Anderluh, M. | | Deposit date: | 2020-05-13 | | Release date: | 2020-11-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent DNA gyrase inhibitors bind asymmetrically to their target using symmetrical bifurcated halogen bonds.
Nat Commun, 12, 2021
|
|
6K35
 
 | | Crystal structure of GH20 exo beta-N-acetylglucosaminidase from Vibrio harveyi in complex with NAG-thiazoline | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, Beta-N-acetylglucosaminidase Nag2 | | Authors: | Meekrathok, P, Stubbs, K.A, Bulmer, D.M, van den Berg, B, Suginta, W. | | Deposit date: | 2019-05-16 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | NAG-thiazoline is a potent inhibitor of the Vibrio campbellii GH20 beta-N-Acetylglucosaminidase.
Febs J., 287, 2020
|
|
5XK2
 
 | |
6ASL
 
 | |
