1ZJ5
 
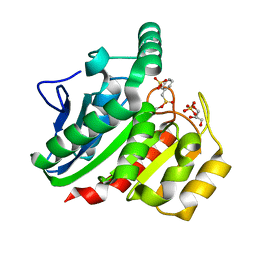 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, C123S, A134S, S208G, A229V, K234R) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
230L
 
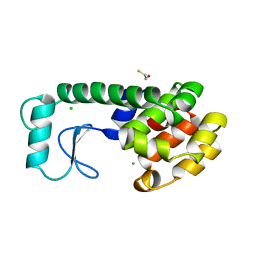 | | T4 LYSOZYME MUTANT M6L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Gassner, N.C, Snow, S, Eldridge, A.M, Drew, D.L, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-02 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
234L
 
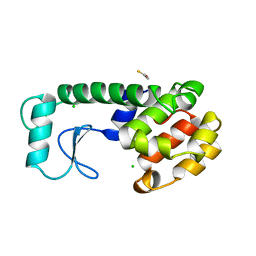 | | T4 LYSOZYME MUTANT M106L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-07 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
2A11
 
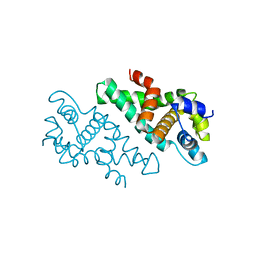 | |
2ADC
 
 | | Solution structure of Polypyrimidine Tract Binding protein RBD34 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
2A07
 
 | | Crystal Structure of Foxp2 bound Specifically to DNA. | | Descriptor: | 5'-D(*AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*T)-3', 5'-D(*TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*G)-3', Forkhead box protein P2, ... | | Authors: | Stroud, J.C, Wu, Y, Bates, D.L, Han, A, Nowick, K, Paabo, S, Tong, H, Chen, L. | | Deposit date: | 2005-06-16 | | Release date: | 2006-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Forkhead Domain of FOXP2 Bound to DNA.
Structure, 14, 2006
|
|
2AD9
 
 | | Solution structure of Polypyrimidine Tract Binding protein RBD1 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
2A2I
 
 | | Aquifex aeolicus KDO8PS in complex with PEP, A5P, Zn2+ | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, ARABINOSE-5-PHOSPHATE, PHOSPHOENOLPYRUVATE, ... | | Authors: | Kona, F, Xu, X, Lu, J, Martin, P, Gatti, D.L. | | Deposit date: | 2005-06-22 | | Release date: | 2006-07-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
232L
 
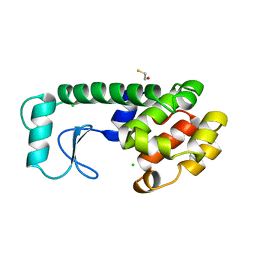 | | T4 LYSOZYME MUTANT M120K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
233L
 
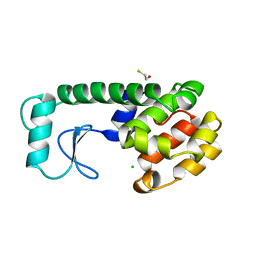 | | T4 LYSOZYME MUTANT M120L | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-07 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
2ADB
 
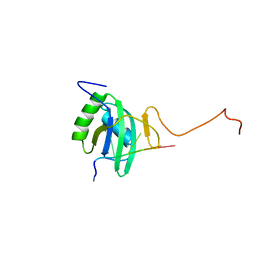 | | Solution structure of Polypyrimidine Tract Binding protein RBD2 complexed with CUCUCU RNA | | Descriptor: | 5'-R(*CP*UP*CP*UP*CP*U)-3', Polypyrimidine tract-binding protein 1 | | Authors: | Oberstrass, F.C, Auweter, S.D, Erat, M, Hargous, Y, Henning, A, Wenter, P, Reymond, L, Pitsch, S, Black, D.L, Allain, F.H.T. | | Deposit date: | 2005-07-20 | | Release date: | 2005-10-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of PTB bound to RNA: specific binding and implications for splicing regulation
Science, 309, 2005
|
|
231L
 
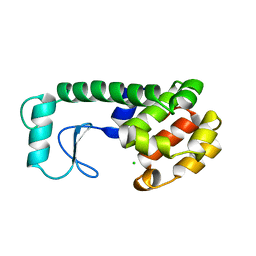 | | T4 LYSOZYME MUTANT M106K | | Descriptor: | CHLORIDE ION, T4 LYSOZYME | | Authors: | Lipscomb, L.A, Drew, D.L, Gassner, N, Baase, W.A, Matthews, B.W. | | Deposit date: | 1997-10-03 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Context-dependent protein stabilization by methionine-to-leucine substitution shown in T4 lysozyme.
Protein Sci., 7, 1998
|
|
1ZIX
 
 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (E36D, R105H, C123S, G211D, K234N)- 1.8 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZHA
 
 | | A. aeolicus KDO8PS R106G mutant in complex with PEP and R5P | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, PHOSPHATE ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-25 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop.
Biochemistry, 44, 2005
|
|
2BSW
 
 | | Crystal structure of a glyphosate-N-acetyltransferase obtained by DNA shuffling. | | Descriptor: | GLYCEROL, GLYPHOSATE N-ACETYLTRANSFERASE, OXIDIZED COENZYME A, ... | | Authors: | Keenan, R.J, Siehl, D.L, Gorton, R, Castle, L.A. | | Deposit date: | 2005-05-24 | | Release date: | 2005-06-08 | | Last modified: | 2015-10-14 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | DNA Shuffling as a Tool for Protein Crystallization.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2A21
 
 | | Aquifex aeolicus KDO8PS in complex with PEP, PO4, and Zn2+ | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, PHOSPHATE ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Kona, F, Xu, X, Lu, J, Martin, P, Gatti, D.L. | | Deposit date: | 2005-06-21 | | Release date: | 2006-06-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Electronic structure of the metal center in the Cd(2+), Zn(2+), and Cu(2+) substituted forms of KDO8P synthase: implications for catalysis.
Biochemistry, 48, 2009
|
|
2B1I
 
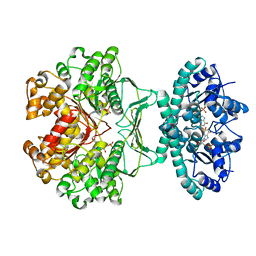 | | crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | Bifunctional purine biosynthesis protein PURH, POTASSIUM ION, [3,4-DIHYDROXY-5R-(2,2,4-TRIOXO-1,2R,3S,4R-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-7-YL)TETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN PHOSPHATE | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
2BKE
 
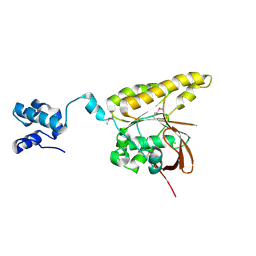 | | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal RadA | | Descriptor: | CHLORIDE ION, DNA REPAIR AND RECOMBINATION PROTEIN RADA | | Authors: | Ariza, A, Richard, D.L, White, M.F, Bond, C.S. | | Deposit date: | 2005-02-15 | | Release date: | 2005-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Conformational Flexibility Revealed by the Crystal Structure of a Crenarchaeal Rada
Nucleic Acids Res., 33, 2005
|
|
1ZI6
 
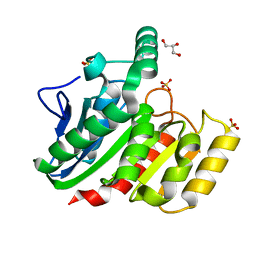 | | Crystal Structure Analysis of the dienelactone hydrolase (C123S) mutant- 1.7 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1ZJI
 
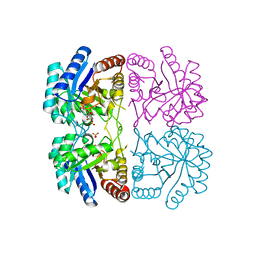 | | Aquifex aeolicus KDO8PS R106G mutant in complex with 2PGA and R5P | | Descriptor: | 2-PHOSPHOGLYCERIC ACID, 2-dehydro-3-deoxyphosphooctonate aldolase, CADMIUM ION, ... | | Authors: | Xu, X, Kona, F, Wang, J, Lu, J, Stemmler, T, Gatti, D.L. | | Deposit date: | 2005-04-28 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Catalytic and Conformational Cycle of Aquifex aeolicus KDO8P Synthase: Role of the L7 Loop
Biochemistry, 44, 2005
|
|
1YXN
 
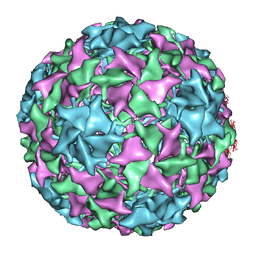 | | Pseudo-atomic model of a fiberless isometric variant of bacteriophage phi29 | | Descriptor: | Major head protein | | Authors: | Morais, M.C, Choi, K.H, Koti, J.S, Chipman, P.R, Anderson, D.L, Rossmann, M.G. | | Deposit date: | 2005-02-22 | | Release date: | 2005-04-26 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Conservation of the Capsid Structure in Tailed dsDNA Bacteriophages: the Pseudoatomic Structure of phi29
Mol.Cell, 18, 2005
|
|
1YYK
 
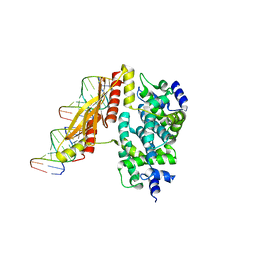 | | Crystal structure of RNase III from Aquifex Aeolicus complexed with double-stranded RNA at 2.5-angstrom resolution | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-R(*CP*GP*CP*GP*AP*AP*UP*UP*CP*GP*CP*G)-3', Ribonuclease III | | Authors: | Gan, J, Tropea, J.E, Austin, B.P, Court, D.L, Waugh, D.S, Ji, X. | | Deposit date: | 2005-02-25 | | Release date: | 2005-11-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intermediate states of ribonuclease III in complex with double-stranded RNA
Structure, 13, 2005
|
|
2BE6
 
 | | 2.0 A crystal structure of the CaV1.2 IQ domain-Ca/CaM complex | | Descriptor: | CALCIUM ION, Calmodulin 2, NICKEL (II) ION, ... | | Authors: | Van Petegem, F, Chatelain, F.C, Minor Jr, D.L. | | Deposit date: | 2005-10-23 | | Release date: | 2005-11-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into voltage-gated calcium channel regulation from the structure of the Ca(V)1.2 IQ domain-Ca(2+)/calmodulin complex
Nat.Struct.Mol.Biol., 12, 2005
|
|
1ZIY
 
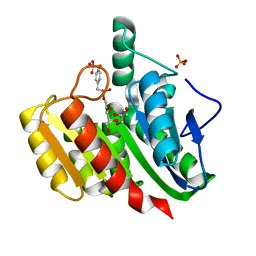 | | Crystal Structure Analysis of the dienelactone hydrolase mutant (C123S) bound with the PMS moiety of the protease inhibitor, Phenylmethylsulfonyl fluoride (PMSF)- 1.9 A | | Descriptor: | Carboxymethylenebutenolidase, GLYCEROL, SULFATE ION | | Authors: | Kim, H.-K, Liu, J.-W, Carr, P.D, Ollis, D.L. | | Deposit date: | 2005-04-27 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Following directed evolution with crystallography: structural changes observed in changing the substrate specificity of dienelactone hydrolase.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2AS5
 
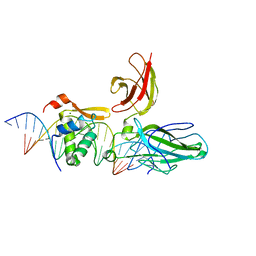 | | Structure of the DNA binding domains of NFAT and FOXP2 bound specifically to DNA. | | Descriptor: | 5'-D(AP*AP*CP*TP*AP*TP*GP*AP*AP*AP*CP*AP*AP*AP*TP*TP*TP*TP*CP*CP*TP*)-3', 5'-D(TP*TP*AP*GP*GP*AP*AP*AP*AP*TP*TP*TP*GP*TP*TP*TP*CP*AP*TP*AP*GP*)-3', Forkhead box protein P2, ... | | Authors: | Wu, Y, Stroud, J.C, Borde, M, Bates, D.L, Guo, L, Han, A, Rao, A, Chen, L. | | Deposit date: | 2005-08-22 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | FOXP3 Controls Regulatory T Cell Function through Cooperation with NFAT.
Cell(Cambridge,Mass.), 126, 2006
|
|
