1MCI
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-D-PHE-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCS
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MEO
 
 | | human glycinamide ribonucleotide Transformylase at pH 4.2 | | Descriptor: | PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase, SULFATE ION | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1MUK
 
 | | reovirus lambda3 native structure | | Descriptor: | MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-24 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1MWH
 
 | | REOVIRUS POLYMERASE LAMBDA3 BOUND TO MRNA CAP ANALOG | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, MANGANESE (II) ION, MINOR CORE PROTEIN LAMBDA 3 | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-09-29 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage-Structural Studies of Reovirus Polymerase lambda3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N38
 
 | | reovirus polymerase lambda3 elongation complex with one phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 3'-DEOXY-URIDINE 5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*C)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
1N35
 
 | | lambda3 elongation complex with four phosphodiester bond formed | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, 5'-R(*AP*UP*UP*AP*GP*CP*CP*CP*CP*C)-3', 5'-R(P*GP*GP*GP*GP*G)-3', ... | | Authors: | Tao, Y, Farsetta, D.L, Nibert, M.L, Harrison, S.C. | | Deposit date: | 2002-10-25 | | Release date: | 2002-12-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Synthesis in a Cage--Structural Studies of Reovirus Polymerase [lambda] 3
Cell(Cambridge,Mass.), 111, 2002
|
|
1KZ8
 
 | | CRYSTAL STRUCTURE OF PORCINE FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH A NOVEL ALLOSTERIC-SITE INHIBITOR | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE MONOPHOSPHATE, FRUCTOSE-1,6-BISPHOSPHATASE, ... | | Authors: | Wright, S.W, Carlo, A.A, Carty, M.D, Danley, D.E, Hageman, D.L, Karam, G.A, Levy, C.B, Mansour, M.N, Mathiowetz, A.M, McClure, L.D, Nestor, N.B, McPherson, R.K, Pandit, J, Pustilnik, L.R, Schulte, G.K, Soeller, W.C, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-02-06 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ANILINOQUINAZOLINE INHIBITORS OF FRUCTOSE 1,6-BISPHOSPHATASE BIND AT A NOVEL ALLOSTERIC SITE: SYNTHESIS, IN VITRO CHARACTERIZATION, AND X-RAY CRYSTALLOGRAPHY
J.MED.CHEM., 45, 2002
|
|
1N71
 
 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii in complex with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Ghuman, N, Wybenga-Groot, L.E, Berghuis, A.M. | | Deposit date: | 2002-11-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the AAC(6')-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members
Protein Sci., 12, 2003
|
|
1LOK
 
 | | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris: A Tale of Buffer Inhibition | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, SODIUM ION, ... | | Authors: | Desmarais, W.T, Bienvenue, D.L, Bzymek, K.P, Holz, R.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The 1.20 Angstrom Resolution Crystal Structure of the Aminopeptidase from Aeromonas proteolytica Complexed with Tris A tale of Buffer Inhibition
Structure, 10, 2002
|
|
4TPL
 
 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OXTOXYNOL-10, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2014-06-08 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1LRN
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1MCC
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-GLN-D-PHE-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1LRO
 
 | | Aquifex aeolicus KDO8P synthase H185G mutant in complex with PEP and Cadmium | | Descriptor: | CADMIUM ION, KDO-8-phosphate synthetase, PHOSPHATE ION, ... | | Authors: | Wang, J, Duewel, H.S, Stuckey, J.A, Woodard, R.W, Gatti, D.L. | | Deposit date: | 2002-05-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Function of His185 in Aquifex aeolicus 3-Deoxy-D-manno-octulosonate 8-Phosphate Synthase
J.Mol.Biol., 324, 2002
|
|
1LEV
 
 | | PORCINE KIDNEY FRUCTOSE-1,6-BISPHOSPHATASE COMPLEXED WITH AN AMP-SITE INHIBITOR | | Descriptor: | 3-(2-CARBOXY-ETHYL)-4,6-DICHLORO-1H-INDOLE-2-CARBOXYLIC ACID, 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, ... | | Authors: | Wright, S.W, Carlo, A.A, Danley, D.E, Hageman, D.L, Karam, G.A, Mansour, M.N, McClure, L.D, Pandit, J, Schulte, G.K, Treadway, J.L, Wang, I.-K, Bauer, P.H. | | Deposit date: | 2002-04-10 | | Release date: | 2002-10-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 3-(2-carboxyethyl)-4,6-dichloro-1H-indole-2-carboxylic acid: an allosteric inhibitor of fructose-1,6-bisphosphatase at the AMP site.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1MCL
 
 | | PRINCIPLES AND PITFALLS in DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), N-ACETYL-D-HIS-L-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MCR
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | IMMUNOGLOBULIN LAMBDA DIMER MCG (LIGHT CHAIN), PEPTIDE N-ACETYL-L-HIS-D-PRO-OH | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1NJS
 
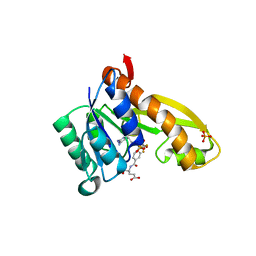 | | human GAR Tfase in complex with hydrolyzed form of 10-trifluoroacetyl-5,10-dideaza-acyclic-5,6,7,8-tetrahydrofolic acid | | Descriptor: | N-{4-[(1R)-4-[(2R,4R,5S)-2,4-DIAMINO-6-OXOHEXAHYDROPYRIMIDIN-5-YL]-1-(2,2,2-TRIFLUORO-1,1-DIHYDROXYETHYL)BUTYL]BENZOYL}-D-GLUTAMIC ACID, PHOSPHATE ION, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Marsilje, T.H, Li, C, Hedrick, M.P, Gooljarsingh, L.T, Tavassoli, A, Benkovic, S.J, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2003-01-02 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Rational Design, Synthesis, Evaluation, and Crystal Structure of a Potent Inhibitor of Human GAR Tfase: 10-(Trifluoroacetyl)-5,10-dideazaacyclic-5,6,7,8-tetrahydrofolic Acid
Biochemistry, 42, 2003
|
|
1NPR
 
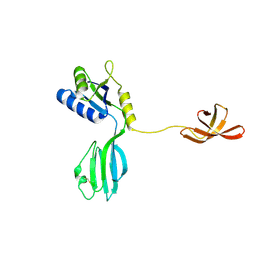 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN C222(1) | | Descriptor: | Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzhan, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
1LCS
 
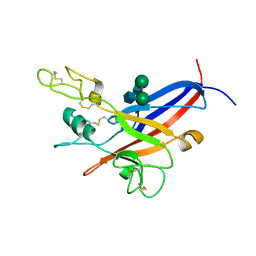 | | RECEPTOR-BINDING DOMAIN FROM SUBGROUP B FELINE LEUKEMIA VIRUS | | Descriptor: | 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Barnett, A.L, Wensel, D.L, Li, W, Fass, D, Cunningham, J.M. | | Deposit date: | 2002-04-06 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Coreceptor for Infection by a pathogenic feline retrovirus
J.Virol., 77, 2003
|
|
1NPP
 
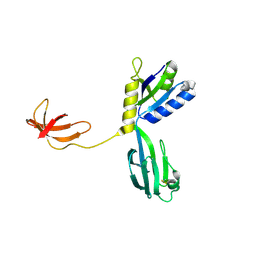 | | CRYSTAL STRUCTURE OF AQUIFEX AEOLICUS NUSG IN P2(1) | | Descriptor: | ISOPROPYL ALCOHOL, Transcription antitermination protein nusG | | Authors: | Knowlton, J.R, Bubunenko, M, Andrykovitch, M, Guo, W, Routzahn, K.M, Waugh, D.S, Court, D.L, Ji, X. | | Deposit date: | 2003-01-18 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Spring-Loaded State of NusG in Its Functional Cycle Is Suggested by X-ray Crystallography and Supported by
Site-Directed Mutants
Biochemistry, 42, 2003
|
|
1N0R
 
 | |
1MCD
 
 | | PRINCIPLES AND PITFALLS IN DESIGNING SITE DIRECTED PEPTIDE LIGANDS | | Descriptor: | Immunoglobulin lambda-1 light chain, PEPTIDE N-ACETYL-D-PHE-B-ALA-L-HIS-D-PRO-NH2 | | Authors: | Edmundson, A.B, Harris, D.L, Fan, Z.-C, Guddat, L.W. | | Deposit date: | 1993-02-25 | | Release date: | 1994-01-31 | | Last modified: | 2020-03-04 | | Method: | X-RAY DIFFRACTION | | Cite: | Principles and pitfalls in designing site-directed peptide ligands.
Proteins, 16, 1993
|
|
1MEN
 
 | | complex structure of human GAR Tfase and substrate beta-GAR | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, Phosphoribosylglycinamide formyltransferase | | Authors: | Zhang, Y, Desharnais, J, Greasley, S.E, Beardsley, G.P, Boger, D.L, Wilson, I.A. | | Deposit date: | 2002-08-08 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Crystal structures of human GAR Tfase of low and high pH and with substrate beta-GAR
Biochemistry, 41, 2002
|
|
1N0Q
 
 | |
