2WBK
 
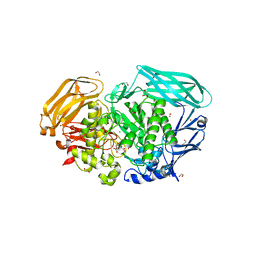 | | Structure of the Michaelis complex of beta-mannosidase, Man2A, provides insight into the conformational itinerary of mannoside hydrolysis | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-mannopyranoside, BETA-MANNOSIDASE, ... | | Authors: | Offen, W.A, Zechel, D.L, Withers, S.G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-03-02 | | Release date: | 2009-03-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Michaelis Complex of Beta-Mannosidase, Man2A, Provides Insight Into the Conformational Itinerary of Mannoside Hydrolysis.
Cell(Cambridge,Mass.), 18, 2009
|
|
2VTF
 
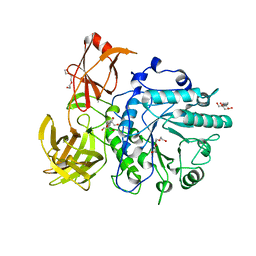 | | X-ray crystal structure of the Endo-beta-N-acetylglucosaminidase from Arthrobacter protophormiae E173Q mutant reveals a TIM barrel catalytic domain and two ancillary domains | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-BETA-N-ACETYLGLUCOSAMINIDASE, TRIETHYLENE GLYCOL | | Authors: | Ling, Z, Bingham, R.J, Suits, M.D.L, Moir, J.W.B, Fairbanks, A.J, Taylor, E.J. | | Deposit date: | 2008-05-14 | | Release date: | 2009-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The X-Ray Crystal Structure of an Arthrobacter Protophormiae Endo-Beta-N-Acetylglucosaminidase Reveals a (Beta/Alpha)(8) Catalytic Domain, Two Ancillary Domains and Active Site Residues Key for Transglycosylation Activity.
J.Mol.Biol., 389, 2009
|
|
1AAW
 
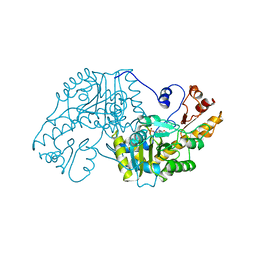 | | THE STRUCTURAL BASIS FOR THE ALTERED SUBSTRATE SPECIFICITY OF THE R292D ACTIVE SITE MUTANT OF ASPARTATE AMINOTRANSFERASE FROM E. COLI | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Almo, S.C, Smith, D.L, Danishefsky, A.T, Ringe, D. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structural basis for the altered substrate specificity of the R292D active site mutant of aspartate aminotransferase from E. coli.
Protein Eng., 7, 1994
|
|
2VOY
 
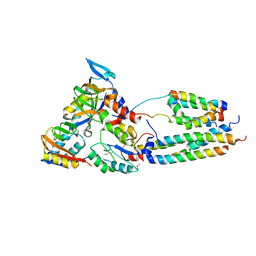 | | CryoEM model of CopA, the copper transporting ATPase from Archaeoglobus fulgidus | | Descriptor: | CATION-TRANSPORTING ATPASE, P-TYPE, POTENTIAL COPPER-TRANSPORTING ATPASE, ... | | Authors: | Wu, C.-C, Rice, W.J, Stokes, D.L. | | Deposit date: | 2008-02-25 | | Release date: | 2009-05-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structure of a Copper Pump Suggests a Regulatory Role for its Metal-Binding Domain.
Structure, 16, 2008
|
|
3HFC
 
 | | A trimeric form of the Kv7.1 A domain Tail, L602M/L606M mutant Semet | | Descriptor: | Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Xu, Q, Minor, D.L. | | Deposit date: | 2009-05-11 | | Release date: | 2009-09-01 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a trimeric form of the K(V)7.1 (KCNQ1) A-domain tail coiled-coil reveals structural plasticity and context dependent changes in a putative coiled-coil trimerization motif.
Protein Sci., 18, 2009
|
|
1A2I
 
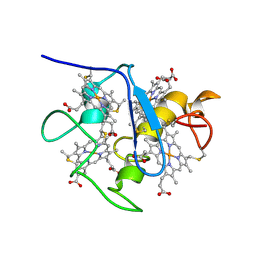 | | SOLUTION STRUCTURE OF DESULFOVIBRIO VULGARIS (HILDENBOROUGH) FERROCYTOCHROME C3, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C3, HEME C | | Authors: | Messias, A.C, Kastrau, D.H.K, Costa, H.S, Legall, J, Turner, D.L, Santos, H, Xavier, A.V. | | Deposit date: | 1998-01-05 | | Release date: | 1998-07-08 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Desulfovibrio vulgaris (Hildenborough) ferrocytochrome c3: structural basis for functional cooperativity.
J.Mol.Biol., 281, 1998
|
|
3HUA
 
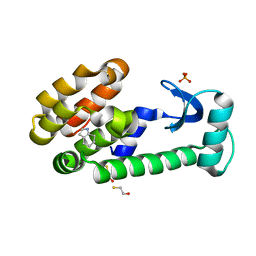 | | 4,5,6,7-tetrahydroindole in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 4,5,6,7-tetrahydro-1H-indole, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
2R1N
 
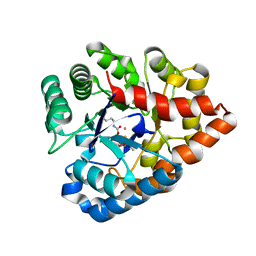 | | OpdA from Agrobacterium radiobacter with bound slow substrate diethyl 4-methoxyphenyl phosphate (20h)- 1.7 A | | Descriptor: | COBALT (II) ION, DIETHYL 4-METHOXYPHENYL PHOSPHATE, FE (II) ION, ... | | Authors: | Ollis, D.L, Jackson, C.J, Foo, J.L, Kim, H.K, Carr, P.D, Liu, J.W, Salem, G. | | Deposit date: | 2007-08-23 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | In crystallo capture of a Michaelis complex and product-binding modes of a bacterial phosphotriesterase
J.Mol.Biol., 375, 2008
|
|
1C3E
 
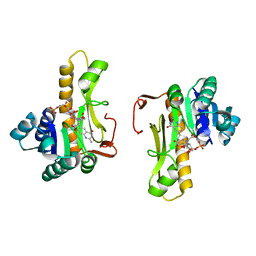 | | NEW INSIGHTS INTO INHIBITOR DESIGN FROM THE CRYSTAL STRUCTURE AND NMR STUDIES OF E. COLI GAR TRANSFORMYLATE IN COMPLEX WITH BETA-GAR AND 10-FORMYL-5,8,10-TRIDEAZAFOLIC ACID. | | Descriptor: | 2-{4-[2-(2-AMINO-4-HYDROXY-QUINAZOLIN-6-YL)-1-CARBOXY-ETHYL]-BENZOYLAMINO}-PENTANEDIOIC ACID, GLYCINAMIDE RIBONUCLEOTIDE, GLYCINAMIDE RIBONUCLEOTIDE TRANSFORMYLASE | | Authors: | Greasley, S.E, Yamashita, M.M, Cai, H, Benkovic, S.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 1999-07-27 | | Release date: | 1999-12-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into inhibitor design from the crystal structure and NMR studies of Escherichia coli GAR transformylase in complex with beta-GAR and 10-formyl-5,8,10-trideazafolic acid.
Biochemistry, 38, 1999
|
|
1CC8
 
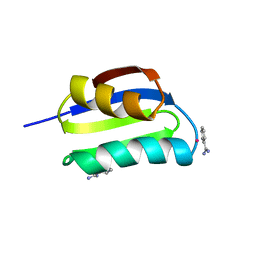 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, MERCURY (II) ION, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
2WVX
 
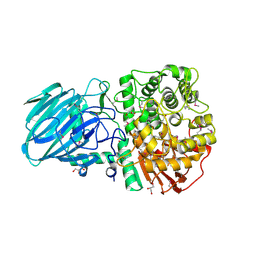 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 | | Descriptor: | CALCIUM ION, GLYCEROL, PUTATIVE ALPHA-1,2-MANNOSIDASE | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WW1
 
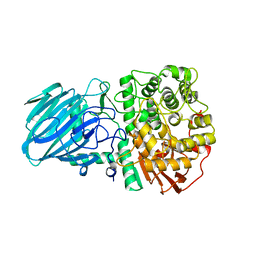 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Thiomannobioside | | Descriptor: | PUTATIVE ALPHA-1,2-MANNOSIDASE, alpha-D-mannopyranose-(1-2)-methyl 2-thio-alpha-D-mannopyranoside | | Authors: | Suits, M.D.L, Zhu, Y, Thompson, A, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-10-21 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
3HU8
 
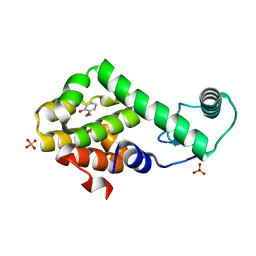 | | 2-ethoxyphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-ethoxyphenol, Lysozyme, PHOSPHATE ION | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-13 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
1C8T
 
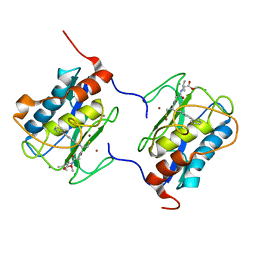 | | HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, el-Kabbani, O, Dunten, P, Crowther, R.L. | | Deposit date: | 1999-07-29 | | Release date: | 2000-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
2RC5
 
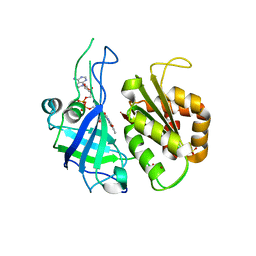 | | Refined structure of FNR from Leptospira interrogans | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, SULFATE ION, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Polikarpov, I, Ceccarelli, E.A. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.431 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
2RC6
 
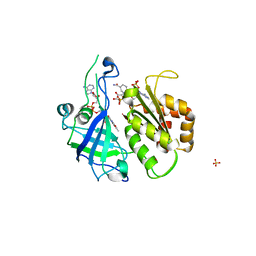 | | Refined structure of FNR from Leptospira interrogans bound to NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin-NADP reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Nascimento, A.S, Catalano-Dupuy, D.L, Ceccarelli, E.A, Polikarpov, I. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Leptospira interrogans FAD-containing ferredoxin-NADP+ reductase and its complex with NADP+.
Bmc Struct.Biol., 7, 2007
|
|
3HTB
 
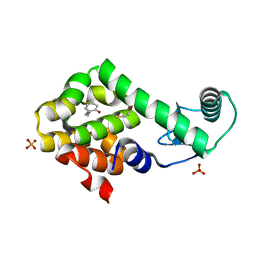 | | 2-propylphenol in complex with T4 lysozyme L99A/M102Q | | Descriptor: | 2-propylphenol, BETA-MERCAPTOETHANOL, Lysozyme, ... | | Authors: | Boyce, S.E, Mobley, D.L, Rocklin, G.J, Graves, A.P, Dill, K.A, Shoichet, B.K. | | Deposit date: | 2009-06-11 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Predicting ligand binding affinity with alchemical free energy methods in a polar model binding site.
J.Mol.Biol., 394, 2009
|
|
1CC7
 
 | | CRYSTAL STRUCTURE OF THE ATX1 METALLOCHAPERONE PROTEIN | | Descriptor: | BENZAMIDINE, PROTEIN (METALLOCHAPERONE ATX1) | | Authors: | Rosenzweig, A.C, Huffman, D.L, Pufahl, M.Y.R.A, Hou, T.V.O, Wernimont, A.K. | | Deposit date: | 1999-03-04 | | Release date: | 1999-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal structure of the Atx1 metallochaperone protein at 1.02 A resolution.
Structure Fold.Des., 7, 1999
|
|
3K84
 
 | | Crystal Structure Analysis of a Oleyl/Oxadiazole/pyridine Inhibitor Bound to a Humanized Variant of Fatty Acid Amide Hydrolase | | Descriptor: | (9Z)-1-(5-pyridin-2-yl-1,3,4-oxadiazol-2-yl)octadec-9-en-1-one, CHLORIDE ION, Fatty-acid amide hydrolase 1 | | Authors: | Mileni, M, Stevens, R.C, Boger, D.L. | | Deposit date: | 2009-10-13 | | Release date: | 2009-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray crystallographic analysis of alpha-ketoheterocycle inhibitors bound to a humanized variant of fatty acid amide hydrolase.
J.Med.Chem., 53, 2010
|
|
2WTE
 
 | | The structure of the CRISPR-associated protein, Csa3, from Sulfolobus solfataricus at 1.8 angstrom resolution. | | Descriptor: | CSA3, DI(HYDROXYETHYL)ETHER | | Authors: | Lintner, N.G, Alsbury, D.L, Copie, V, Young, M.J, Lawrence, C.M. | | Deposit date: | 2009-09-15 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Structure of the Crispr-Associated Protein Csa3 Provides Insight Into the Regulation of the Crispr/Cas System.
J.Mol.Biol., 405, 2011
|
|
2WHJ
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, GLYCEROL, ... | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
3J1Z
 
 | | Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy | | Descriptor: | Cation efflux family protein | | Authors: | Coudray, N, Valvo, S, Hu, M, Lasala, R, Kim, C, Vink, M, Zhou, M, Provasi, D, Filizola, M, Tao, J, Fang, J, Penczek, P.A, Ubarretxena-Belandia, I, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2WJ2
 
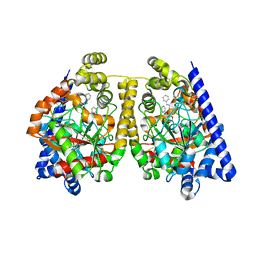 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(5-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(5-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
1APH
 
 | | CONFORMATIONAL CHANGES IN CUBIC INSULIN CRYSTALS IN THE PH RANGE 7-11 | | Descriptor: | 1,2-DICHLOROETHANE, INSULIN A CHAIN (PH 7), INSULIN B CHAIN (PH 7) | | Authors: | Gursky, O, Badger, J, Li, Y, Caspar, D.L.D. | | Deposit date: | 1992-10-30 | | Release date: | 1993-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational changes in cubic insulin crystals in the pH range 7-11.
Biophys.J., 63, 1992
|
|
2WJ1
 
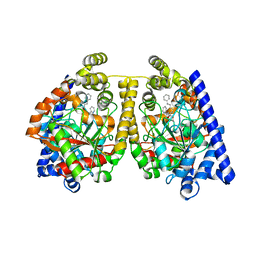 | | 3D-crystal structure of humanized-rat fatty acid amide hydrolase (FAAH) conjugated with 7-phenyl-1-(4-(pyridin-2-yl)oxazol-2-yl)heptan- 1-one, an alpha-ketooxazole | | Descriptor: | 7-phenyl-1-(4-pyridin-2-yl-1,3-oxazol-2-yl)heptane-1,1-diol, CHLORIDE ION, FATTY-ACID AMIDE HYDROLASE 1 | | Authors: | Mileni, M, Garfunkle, J, DeMartino, J.K, Cravatt, B.F, Boger, D.L, Stevens, R.C. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Binding and Inactivation Mechanism of a Humanized Fatty Acid Amide Hydrolase by Alpha-Ketoheterocycle Inhibitors Revealed from Cocrystal Structures.
J.Am.Chem.Soc., 131, 2009
|
|
