3T9L
 
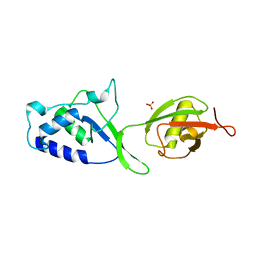 | | Structure of N-terminal DUSP-UBL domains of human USP15 | | Descriptor: | SULFATE ION, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Harper, S, Besong, T.M.D, Emsley, J, Scott, D.J, Dreveny, I. | | Deposit date: | 2011-08-03 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the USP15 N-Terminal Domains: A beta-Hairpin Mediates Close Association between the DUSP and UBL Domains
Biochemistry, 50, 2011
|
|
4JS7
 
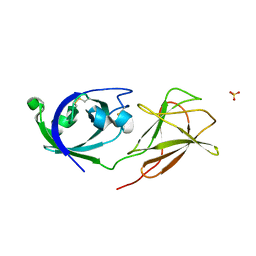 | |
3TFJ
 
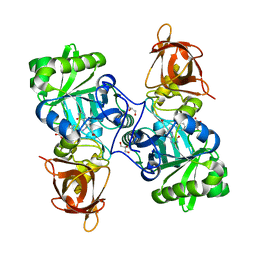 | | DMSP-dependent demethylase from P. ubique - with cofactor THF | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, ACETATE ION, GLYCEROL, ... | | Authors: | Schuller, D.J, Reisch, C.R, Moran, M.A, Whitman, W.B, Lanzilotta, W.N. | | Deposit date: | 2011-08-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of dimethylsulfoniopropionate-dependent demethylase from the marine organism Pelagabacter ubique.
Protein Sci., 21, 2012
|
|
4H9P
 
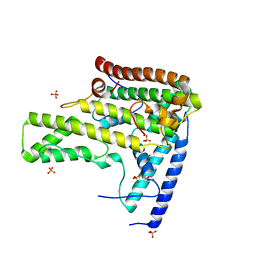 | | Complex structure 3 of DAXX/H3.3(sub5,G90A)/H4 | | Descriptor: | Death domain-associated protein 6, Histone H3.3, Histone H4, ... | | Authors: | Elsasser, S.J, Huang, H, Lewis, P.W, Chin, J.W, Allis, D.C, Patel, D.J. | | Deposit date: | 2012-09-24 | | Release date: | 2012-10-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | DAXX chaperone envelops an H3.3/H4 dimer dictating H3.3-specific read out
To be Published
|
|
3S6P
 
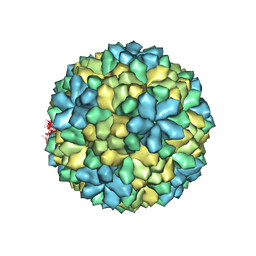 | |
3SKT
 
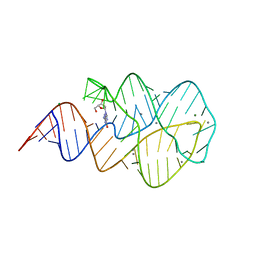 | | Crystal structure of the 2'- Deoxyguanosine riboswitch bound to 2'- Deoxyguanosine, manganese Soak | | Descriptor: | 2'-DEOXY-GUANOSINE, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Pikovskaya, O, Polonskaia, A, Patel, D.J, Serganov, A. | | Deposit date: | 2011-06-23 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural principles of nucleoside selectivity in a 2'-deoxyguanosine riboswitch.
Nat.Chem.Biol., 7, 2011
|
|
4HM1
 
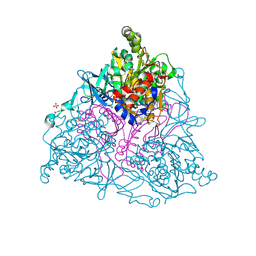 | | Naphthalene 1,2-Dioxygenase bound to 1-indanone | | Descriptor: | 1,2-ETHANEDIOL, 2,3-dihydro-1H-inden-1-one, FE (III) ION, ... | | Authors: | Ferraro, D.J, Ramaswamy, S. | | Deposit date: | 2012-10-17 | | Release date: | 2013-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Naphthalene 1,2-Dioxygenase bound to 1-indanone
To be Published
|
|
3RV0
 
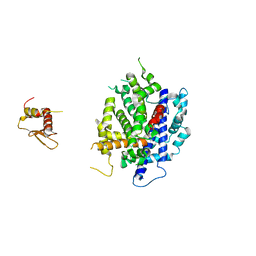 | | Crystal structure of K. polysporus Dcr1 without the C-terminal dsRBD | | Descriptor: | K. polysporus Dcr1, MAGNESIUM ION | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2011-05-05 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The inside-out mechanism of dicers from budding yeasts.
Cell(Cambridge,Mass.), 146, 2011
|
|
4JGZ
 
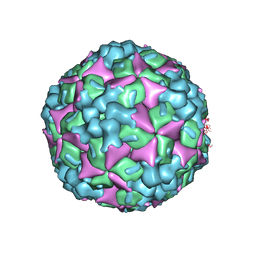 | | Crystal structure of human coxsackievirus A16 uncoating intermediate (space group I222) | | Descriptor: | Polyprotein, capsid protein VP1, capsid protein VP2, ... | | Authors: | Ren, J, Wang, X, Hu, Z, Gao, Q, Sun, Y, Li, X, Porta, C, Walter, T.S, Gilbert, R.J, Zhao, Y, Axford, D, Williams, M, McAuley, K, Rowlands, D.J, Yin, W, Wang, J, Stuart, D.I, Rao, Z, Fry, E.E. | | Deposit date: | 2013-03-04 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Picornavirus uncoating intermediate captured in atomic detail.
Nat Commun, 4, 2013
|
|
4JQ5
 
 | |
3RZN
 
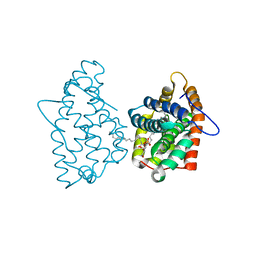 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with 3-O-sulfo-galactosylceramide containing nervonoyl acyl chain (24:1) | | Descriptor: | (15Z)-N-((1S,2R,3E)-2-HYDROXY-1-{[(3-O-SULFO-BETA-D-GALACTOPYRANOSYL)OXY]METHYL}HEPTADEC-3-ENYL)TETRACOS-15-ENAMIDE, Glycolipid transfer protein | | Authors: | Samygina, V, Cabo-Bilbao, A, Popov, A.N, Ochoa-Lizarralde, B, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-12 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
3PMQ
 
 | |
4I4E
 
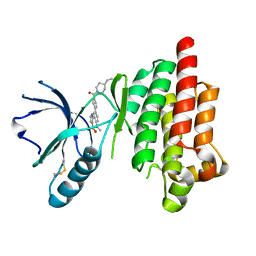 | | Structure of Focal Adhesion Kinase catalytic domain in complex with hinge binding pyrazolobenzothiazine compound. | | Descriptor: | Focal adhesion kinase 1, [4-(2-hydroxyethyl)piperidin-1-yl][4-(5-methyl-4,4-dioxido-1,5-dihydropyrazolo[4,3-c][2,1]benzothiazin-8-yl)phenyl]methanone | | Authors: | Skene, R.J, Hosfield, D.J. | | Deposit date: | 2012-11-27 | | Release date: | 2013-03-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-based discovery of cellular-active allosteric inhibitors of FAK.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3Q0B
 
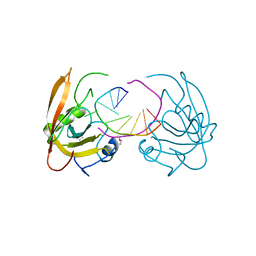 | |
3Q0F
 
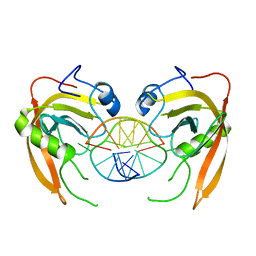 | | Crystal structure of SUVH5 SRA- methylated CHH DNA complex | | Descriptor: | DNA (5'-D(*CP*TP*GP*AP*GP*GP*AP*GP*TP*AP*T)-3'), DNA (5'-D(*TP*AP*CP*TP*(5CM)P*CP*TP*CP*AP*G)-3'), Histone-lysine N-methyltransferase, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
4HZ3
 
 | | MthK pore crystallized in presence of TBSb | | Descriptor: | Calcium-gated potassium channel mthK, HEXANE-1,6-DIOL, POTASSIUM ION | | Authors: | Posson, D.J, McCoy, J.G, Nimigean, C.M. | | Deposit date: | 2012-11-14 | | Release date: | 2012-12-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The voltage-dependent gate in MthK potassium channels is located at the selectivity filter.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3Q0C
 
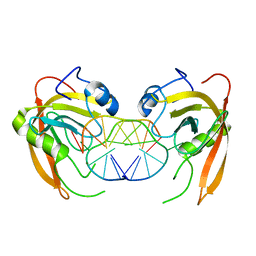 | | Crystal structure of SUVH5 SRA-fully methylated CG DNA complex in space group P6122 | | Descriptor: | DNA (5'-D(*AP*CP*TP*AP*(5CM)P*GP*TP*AP*GP*TP*T)-3'), Histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH5, ... | | Authors: | Eerappa, R, Simanshu, D.K, Patel, D.J. | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6567 Å) | | Cite: | A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo.
Genes Dev., 25, 2011
|
|
3U7U
 
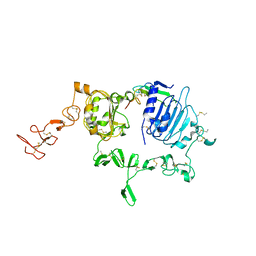 | | Crystal structure of extracellular region of human epidermal growth factor receptor 4 in complex with neuregulin-1 beta | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuregulin 1, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Liu, P, Cleveland IV, T.E, Bouyain, S, Longo, P.A, Leahy, D.J. | | Deposit date: | 2011-10-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | A single ligand is sufficient to activate EGFR dimers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4JOL
 
 | |
4JNV
 
 | | Crystal structure of the human Nup57CCS3* coiled-coil segment, space group C2 | | Descriptor: | Nucleoporin p54 | | Authors: | Stuwe, T, Bley, C.J, Mayo, D.J, Hoelz, A. | | Deposit date: | 2013-03-15 | | Release date: | 2014-09-17 | | Last modified: | 2016-02-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Architecture of the fungal nuclear pore inner ring complex.
Science, 350, 2015
|
|
3QRY
 
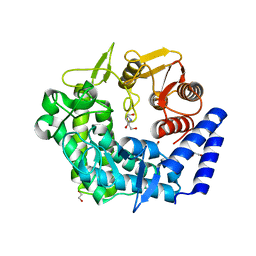 | | Analysis of a new family of widely distributed metal-independent alpha mannosidases provides unique insight into the processing of N-linked glycans, Streptococcus pneumoniae SP_2144 1-deoxymannojirimycin complex | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYMANNOJIRIMYCIN, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2011-02-18 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
4IOY
 
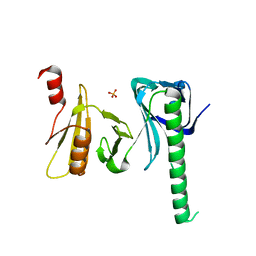 | |
3QHB
 
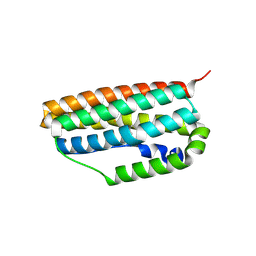 | |
3QOQ
 
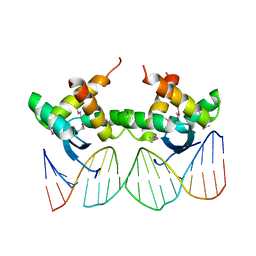 | |
3QPF
 
 | | Analysis of a New Family of Widely Distributed Metal-independent alpha-Mannosidases Provides Unique Insight into the Processing of N-linked Glycans, Streptococcus pneumoniae SP_2144 apo-structure | | Descriptor: | 1,2-ETHANEDIOL, Putative uncharacterized protein | | Authors: | Gregg, K.J, Zandberg, W.F, Hehemann, J.-H, Whitworth, G.E, Deng, L.E, Vocadlo, D.J. | | Deposit date: | 2011-02-12 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Analysis of a New Family of Widely Distributed Metal-independent {alpha}-Mannosidases Provides Unique Insight into the Processing of N-Linked Glycans.
J.Biol.Chem., 286, 2011
|
|
