7MEJ
 
 | |
5J7X
 
 | | Baeyer-Villiger monooxygenase BVMOAFL838 from Aspergillus flavus | | 分子名称: | Dimethylaniline monooxygenase, putative, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Ferroni, F.M, Tolmie, C, Smit, M.S, Opperman, D.J. | | 登録日 | 2016-04-07 | | 公開日 | 2016-08-10 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Catalytic Characterization of a Fungal Baeyer-Villiger Monooxygenase.
Plos One, 11, 2016
|
|
7MDW
 
 | |
7ME7
 
 | |
2B5W
 
 | | Crystal structure of D38C glucose dehydrogenase mutant from Haloferax mediterranei | | 分子名称: | CITRATE ANION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | 著者 | Britton, K.L, Baker, P.J, Fisher, M, Ruzheinikov, S, Gilmour, D.J, Bonete, M.-J, Ferrer, J, Pire, C, Esclapez, J, Rice, D.W. | | 登録日 | 2005-09-29 | | 公開日 | 2006-04-04 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Analysis of protein solvent interactions in glucose dehydrogenase from the extreme halophile Haloferax mediterranei.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2Y9E
 
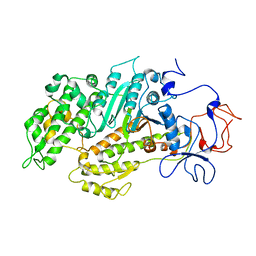 | | Structural basis for the allosteric interference of myosin function by mutants G680A and G680V of Dictyostelium myosin-2 | | 分子名称: | MYOSIN-2 | | 著者 | Preller, M, Bauer, S, Adamek, N, Fujita-Becker, S, Fedorov, R, Geeves, M.A, Manstein, D.J. | | 登録日 | 2011-02-14 | | 公開日 | 2011-07-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.397 Å) | | 主引用文献 | Structural Basis for the Allosteric Interference of Myosin Function by Reactive Thiol Region Mutations G680A and G680V.
J.Biol.Chem., 286, 2011
|
|
5FQH
 
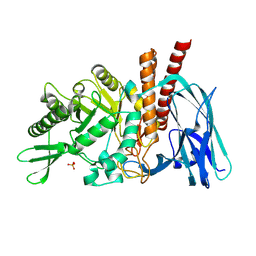 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-4)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | 著者 | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | 登録日 | 2015-12-10 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
7NIT
 
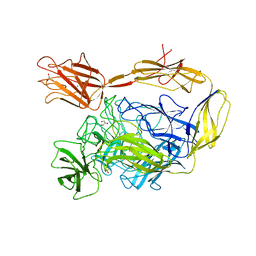 | | X-ray structure of a multidomain BbgIII from Bifidobacterium bifidum | | 分子名称: | Beta-galactosidase, CALCIUM ION, GLYCEROL, ... | | 著者 | Moroz, O.V, Blagova, E, Lebedev, A.A, Sanchez Rodriguez, F, Rigden, D.J, Tams, J.W, Wilting, R, Vester, J.K, Longhin, E, Krogh, K.B.R, Pache, R.A, Davies, G.J, Wilson, K.S. | | 登録日 | 2021-02-14 | | 公開日 | 2021-12-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Multitasking in the gut: the X-ray structure of the multidomain BbgIII from Bifidobacterium bifidum offers possible explanations for its alternative functions.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
2XZS
 
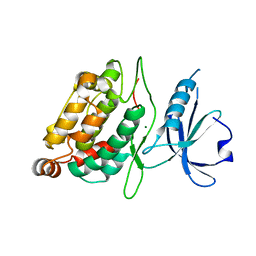 | | Death associated protein kinase 1 residues 1-312 | | 分子名称: | DEATH ASSOCIATED KINASE 1, MAGNESIUM ION | | 著者 | Yumerefendi, H, Mas, P.J, Dordevic, N, McCarthy, A.A, Hart, D.J. | | 登録日 | 2010-11-29 | | 公開日 | 2011-12-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Death-Associated Protein Kinase Activity is Regulated by Coupled Calcium/Calmodulin Binding to Two Distinct Sites.
Structure, 24, 2016
|
|
5FK0
 
 | |
5FR0
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | 分子名称: | 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, PHOSPHATE ION | | 著者 | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | 登録日 | 2015-12-14 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
2BGZ
 
 | | ATOMIC MODEL OF THE BACTERIAL FLAGELLAR BASED ON DOCKING AN X-RAY DERIVED HOOK STRUCTURE INTO AN EM MAP. | | 分子名称: | FLAGELLAR HOOK PROTEIN FLGE | | 著者 | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, Derosier, D.J. | | 登録日 | 2005-01-06 | | 公開日 | 2005-01-27 | | 最終更新日 | 2024-05-08 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
5ER5
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC1990 | | 分子名称: | CALCIUM ION, ETHIDIUM, Protein S100-B | | 著者 | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | 登録日 | 2015-11-13 | | 公開日 | 2016-06-08 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.26 Å) | | 主引用文献 | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
2XX0
 
 | | STRUCTURE OF THE N90S-H254F MUTANT OF NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | 著者 | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | 登録日 | 2010-11-07 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
5HPN
 
 | |
7LHC
 
 | |
5DR6
 
 | | Aurora A Kinase in Complex with AA30 and JNJ-7706621 in Space Group P6122 | | 分子名称: | 2-(3-bromophenyl)quinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A | | 著者 | Janecek, M, Rossmann, M, Sharma, P, Emery, A, McKenzie, G.J, Huggins, D.J, Stockwell, S, Stokes, J.A, Almeida, E.G, Hardwick, B, Narvaez, A.J, Hyvonen, M, Spring, D.R, Venkitaraman, A.R. | | 登録日 | 2015-09-15 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.534 Å) | | 主引用文献 | Allosteric modulation of AURKA kinase activity by a small-molecule inhibitor of its protein-protein interaction with TPX2.
Sci Rep, 6, 2016
|
|
2XWZ
 
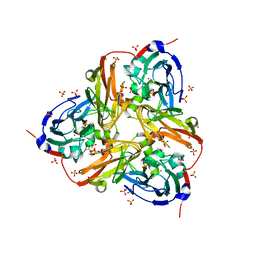 | | STRUCTURE OF THE RECOMBINANT NATIVE NITRITE REDUCTASE FROM ALCALIGENES XYLOSOXIDANS complexed with nitrite | | 分子名称: | ACETATE ION, COPPER (II) ION, DISSIMILATORY COPPER-CONTAINING NITRITE REDUCTASE, ... | | 著者 | Antonyuk, S.V, Leferink, N.G.H, Han, C, Heyes, D.J, Rigby, S.E.J, Hough, M.A, Eady, R.R, Scrutton, N.S, Hasnain, S.S. | | 登録日 | 2010-11-06 | | 公開日 | 2011-05-18 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Proton-Coupled Electron Transfer in the Catalytic Cycle of Alcaligenes Xylosoxidans Copper-Dependent Nitrite Reductase.
Biochemistry, 50, 2011
|
|
5DUN
 
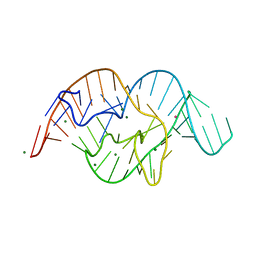 | | The crystal structure of OMe substituted twister ribozyme | | 分子名称: | MAGNESIUM ION, POTASSIUM ION, RNA (54-MER) | | 著者 | Ren, A, Patel, D.J, Micura, R, Rajashankar, K.R. | | 登録日 | 2015-09-19 | | 公開日 | 2015-10-28 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.64 Å) | | 主引用文献 | A Mini-Twister Variant and Impact of Residues/Cations on the Phosphodiester Cleavage of this Ribozyme Class.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2Y0A
 
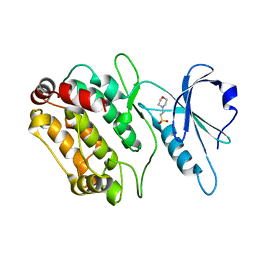 | | Structure of DAPK1 construct residues 1-304 | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DEATH-ASSOCIATED PROTEIN KINASE 1 | | 著者 | Yumerefendi, H, Mas, P.J, Dordevic, N, McCarthy, A.A, Hart, D.J. | | 登録日 | 2010-12-01 | | 公開日 | 2011-12-14 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Library-Based Construct Screening of Death-Associated Protein Kinase 1 Identifies the Minimal Calmodulin Interaction Region and Autoinhibitory Conformation of the Catalytic Domain
To be Published
|
|
5JDX
 
 | |
5FQF
 
 | | The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens | | 分子名称: | 2-acetamido-2-deoxy-beta-D-galactopyranose, BETA-N-ACETYLGALACTOSAMINIDASE, FORMIC ACID | | 著者 | Noach, I, Pluvinage, B, Laurie, C, Abe, K.T, Alteen, M, Vocadlo, D.J, Boraston, A.B. | | 登録日 | 2015-12-10 | | 公開日 | 2016-03-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The Details of Glycolipid Glycan Hydrolysis by the Structural Analysis of a Family 123 Glycoside Hydrolase from Clostridium Perfringens
J.Mol.Biol., 428, 2016
|
|
2YA0
 
 | | Catalytic Module of the Multi-modular glycogen-degrading pneumococcal virulence factor SpuA | | 分子名称: | CALCIUM ION, GLYCEROL, PUTATIVE ALKALINE AMYLOPULLULANASE, ... | | 著者 | Lammerts van Bueren, A, Ficko-Blean, E, Pluvinage, B, Hehemann, J.H, Higgins, M.A, Deng, L, Ogunniyi, A.D, Stroeher, U.H, Warry, N.E, Burke, R.D, Czjzek, M, Paton, J.C, Vocadlo, D.J, Boraston, A.B. | | 登録日 | 2011-02-17 | | 公開日 | 2011-04-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | The Conformation and Function of a Multimodular Glycogen-Degrading Pneumococcal Virulence Factor.
Structure, 19, 2011
|
|
5FL1
 
 | | Structure of a hydrolase with an inhibitor | | 分子名称: | (3~{a}~{R},5~{R},6~{S},7~{R},7~{a}~{R})-5-(hydroxymethyl)-2-(prop-2-enylamino)-5,6,7,7~{a}-tetrahydro-3~{a}~{H}-pyrano[3,2-d][1,3]thiazole-6,7-diol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | 著者 | Cekic, N, Heinonen, J.E, Stubbs, K.A, Roth, C, McEachern, E.J, Davies, G.J, Vocadlo, D.J. | | 登録日 | 2015-10-20 | | 公開日 | 2016-08-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Analysis of transition state mimicry by tight binding aminothiazoline inhibitors provides insight into catalysis by humanO-GlcNAcase.
Chem Sci, 7, 2016
|
|
2YNN
 
 | | yeast betaprime COP 1-304 with KTKTN motif | | 分子名称: | COATOMER SUBUNIT BETA', KTKTN MOTIF, SULFATE ION | | 著者 | Jackson, L.P, Lewis, M, Kent, H.M, Edeling, M.A, Evans, P.R, Duden, R, Owen, D.J. | | 登録日 | 2012-10-17 | | 公開日 | 2012-12-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.782 Å) | | 主引用文献 | Molecular Basis for Recognition of Dilysine Trafficking Motifs by Copi.
Dev.Cell, 23, 2012
|
|
