4EZ4
 
 | | free KDM6B structure | | Descriptor: | Lysine-specific demethylase 6B, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Cheng, Z.J, Patel, D.J. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Nature, 488, 2012
|
|
4F9D
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with nickel | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
4F9J
 
 | | Structure of Escherichia coli PgaB 42-655 in complex with iron | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CALCIUM ION, ... | | Authors: | Little, D.J, Poloczek, J, Whitney, J.C, Robinson, H, Nitz, M, Howell, P.L. | | Deposit date: | 2012-05-18 | | Release date: | 2012-07-25 | | Last modified: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | The Structure and Metal Dependent Activity of Escherichia coli PgaB Provides Insight into the Partial De-N-acetylation of Poly-b-1,6-N-acetyl-D-glucosamine
J.Biol.Chem., 287, 2012
|
|
4EYU
 
 | |
4F1N
 
 | | Crystal structure of Kluyveromyces polysporus Argonaute with a guide RNA | | Descriptor: | KpAGO, RNA 5'-R(P*UP*AP*AP*AP*AP*AP*AP*AP*A)-3' | | Authors: | Nakanishi, K, Weinberg, D.E, Bartel, D.P, Patel, D.J. | | Deposit date: | 2012-05-07 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.187 Å) | | Cite: | Structure of yeast Argonaute with guide RNA.
Nature, 486, 2012
|
|
4FG2
 
 | |
4FG4
 
 | |
4FQO
 
 | | Crystal Structure of Calcium-Loaded S100B Bound to SBi4211 | | Descriptor: | 4,4'-[heptane-1,7-diylbis(oxy)]dibenzenecarboximidamide, CALCIUM ION, Protein S100-B | | Authors: | McKnight, L.E, Raman, E.P, Bezawada, P, Kudrimoti, S, Wilder, P.T, Hartman, K.G, Toth, E.A, Coop, A, MacKerrell, A.D, Weber, D.J. | | Deposit date: | 2012-06-25 | | Release date: | 2012-10-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Based Discovery of a Novel Pentamidine-Related Inhibitor of the Calcium-Binding Protein S100B.
ACS Med Chem Lett, 3, 2012
|
|
2X8U
 
 | | Sphingomonas wittichii Serine palmitoyltransferase | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Raman, M.C.C, Johnson, K.A, Campopiano, D.J, Naismith, J.H. | | Deposit date: | 2010-03-12 | | Release date: | 2010-03-23 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Serine Palmitoyltransferase from Sphingomonas Wittichii Rw1 an Interesting Link to an Unusual Acyl Carrier Protein
Biopolymers, 93, 2010
|
|
4TZ0
 
 | | DEAD-box helicase Mss116 bound to ssRNA and GDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TYW
 
 | | DEAD-box helicase Mss116 bound to ssRNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.197 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TZ6
 
 | | DEAD-box helicase Mss116 bound to ssRNA and UDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TYY
 
 | | DEAD-box helicase Mss116 bound to ssRNA and CDP-BeF | | Descriptor: | ATP-dependent RNA helicase MSS116, mitochondrial, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-09 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4TYN
 
 | | DEAD-box helicase Mss116 bound to ssDNA and ADP-BeF | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent RNA helicase MSS116, mitochondrial, ... | | Authors: | Mallam, A.L, Sidote, D.J, Lambowitz, A.M. | | Deposit date: | 2014-07-08 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.959 Å) | | Cite: | Molecular insights into RNA and DNA helicase evolution from the determinants of specificity for a DEAD-box RNA helicase.
Elife, 3, 2014
|
|
4UN1
 
 | | Sirohaem decarboxylase AhbA/B - an enzyme with structural homology to the Lrp/AsnC transcription factor family that is part of the alternative haem biosynthesis pathway. | | Descriptor: | 12,18-DIDECARBOXY-SIROHEME, PUTATIVE TRANSCRIPTIONAL REGULATOR, ASNC FAMILY | | Authors: | Palmer, D.J, Brown, D.G, Warren, M.J, Pickersgill, R.W. | | Deposit date: | 2014-05-23 | | Release date: | 2014-06-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structure, Function and Properties of Sirohaem Decarboxylase - an Enzyme with Structural Homology to a Transcription Factor Family that is Part of the Alternative Haem Biosynthesis Pathway.
Mol.Microbiol., 93, 2014
|
|
4W7F
 
 | |
4V7H
 
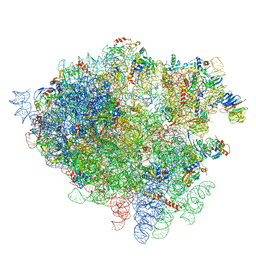 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
4W74
 
 | |
3U3F
 
 | | Structural basis for the interaction of Pyk2 PAT domain with paxillin LD motifs | | Descriptor: | Paxillin LD2 peptide, Protein-tyrosine kinase 2-beta | | Authors: | Vanarotti, M, Miller, D.J, Guibao, C.C, Zheng, J.J. | | Deposit date: | 2011-10-05 | | Release date: | 2012-10-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | Structural and Mechanistic Insights into the Interaction between Pyk2 and Paxillin LD Motifs.
J.Mol.Biol., 426, 2014
|
|
3KHR
 
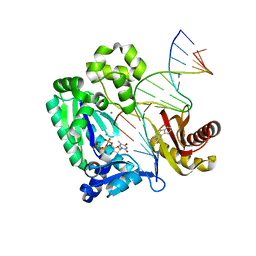 | | Dpo4 post-extension ternary complex with the correct C opposite the 2-aminofluorene-guanine [AF]G lesion | | Descriptor: | 2-AMINOFLUORENE, 5'-D(*CP*C*TP*AP*AP*CP*GP*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*GP*CP*(DDG))-3', ... | | Authors: | Rechkoblit, O, Malinina, L, Patel, D.J. | | Deposit date: | 2009-10-30 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mechanism of error-free and semitargeted mutagenic bypass of an aromatic amine lesion by Y-family polymerase Dpo4.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3U5P
 
 | |
3KT1
 
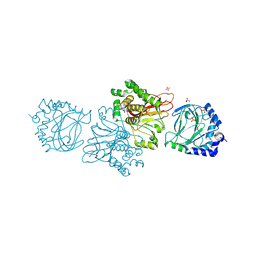 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, GLYCEROL, PKHD-type hydroxylase TPA1, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3U9U
 
 | | Crystal Structure of Extracellular Domain of Human ErbB4/Her4 in complex with the Fab fragment of mAb1479 | | Descriptor: | Fab Heavy Chain, Fab Light Chain, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Hollmen, M, Liu, P, Wildiers, H, Reinvall, I, Vandorpe, T, Smeets, A, Deraedt, K, Vahlberg, T, Joensuu, H, Leahy, D.J, Schoffski, P, Elenius, K. | | Deposit date: | 2011-10-19 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Proteolytic processing of ErbB4 in breast cancer.
Plos One, 7, 2012
|
|
3U5N
 
 | |
3KT7
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
