1KQI
 
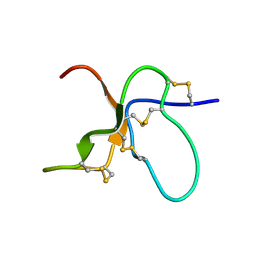 | | NMR Solution Structure of the trans Pro30 Isomer of ACTX-Hi:OB4219 | | Descriptor: | ACTX-Hi:OB4219 | | Authors: | Rosengren, K.J, Wilson, D, Daly, N.L, Alewood, P.F, Craik, D.J. | | Deposit date: | 2002-01-06 | | Release date: | 2002-02-06 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the cis- and trans-Pro30 isomers of a novel 38-residue toxin
from the venom of Hadronyche Infensa sp. that contains a cystine-knot motif within
its four disulfide bonds
Biochemistry, 41, 2002
|
|
2JCP
 
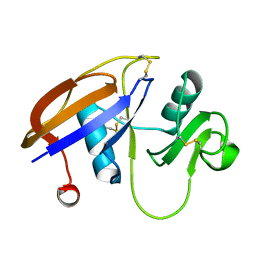 | | The hyaluronan binding domain of murine CD44 | | Descriptor: | CD44 ANTIGEN | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2K14
 
 | | Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis | | Descriptor: | YuaF protein | | Authors: | Walker, C.A, Hinderhofer, M, Witte, D.J, Boos, W, Moller, H.M. | | Deposit date: | 2008-02-20 | | Release date: | 2008-08-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the soluble domain of the NfeD protein YuaF from Bacillus subtilis.
J.Biomol.Nmr, 42, 2008
|
|
2JSM
 
 | | MONOMERIC HUMAN TELOMERE DNA TETRAPLEX WITH 3+1 STRAND FOLD TOPOLOGY, TWO EDGEWISE LOOPS AND DOUBLE-CHAIN REVERSAL LOOP, NMR, 10 STRUCTURES, Form 1 Natural | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-09 | | Release date: | 2008-07-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution
Nucleic Acids Res., 35, 2007
|
|
2HT7
 
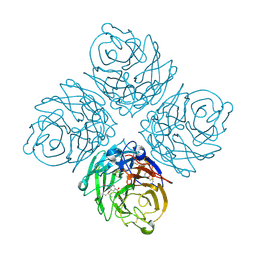 | | N8 neuraminidase in open complex with oseltamivir | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-25 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2HTU
 
 | | N8 neuraminidase in complex with peramivir | | Descriptor: | 3-(1-ACETYLAMINO-2-ETHYL-BUTYL)-4-GUANIDINO-2-HYDROXY-CYCLOPENTANECARBOXYLIC ACID, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2HMN
 
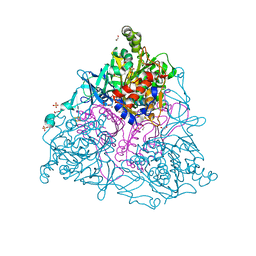 | | Crystal Structure of the Naphthalene 1,2-Dioxygenase F352V Mutant Bound to Anthracene. | | Descriptor: | 1,2-ETHANEDIOL, ANTHRACENE, FE (III) ION, ... | | Authors: | Ferraro, D.J, Okerlund, A.L, Mowers, J.C, Ramaswamy, S. | | Deposit date: | 2006-07-11 | | Release date: | 2006-10-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for regioselectivity and stereoselectivity of product formation by naphthalene 1,2-dioxygenase.
J.Bacteriol., 188, 2006
|
|
2HTW
 
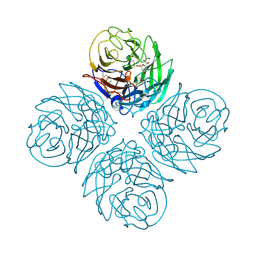 | | N4 neuraminidase in complex with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Neuraminidase | | Authors: | Russell, R.J, Haire, L.F, Stevens, D.J, Collins, P.J, Lin, Y.P, Blackburn, G.M, Hay, A.J, Gamblin, S.J, Skehel, J.J. | | Deposit date: | 2006-07-26 | | Release date: | 2006-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design.
Nature, 443, 2006
|
|
2JMI
 
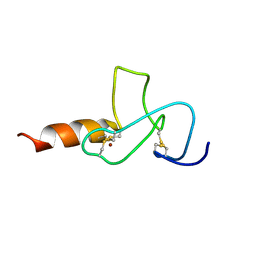 | | NMR solution structure of PHD finger fragment of Yeast Yng1 protein in free state | | Descriptor: | Protein YNG1, ZINC ION | | Authors: | Ilin, S, Taverna, S.D, Rogers, R.S, Tanny, J.C, Lavender, H, Li, H, Baker, L, Boyle, J, Blair, L.P, Chait, B.T, Patel, D.J, Aitchison, J.D, Tackett, A.J, Allis, C.D. | | Deposit date: | 2006-11-15 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs
Mol.Cell, 24, 2006
|
|
2JCR
 
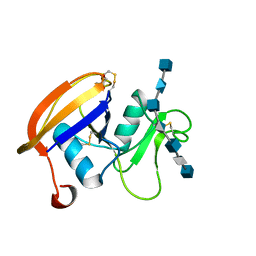 | | The hyaluronan binding domain of murine CD44 in a Type B complex with an HA 8-mer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD44 ANTIGEN, GLYCEROL | | Authors: | Banerji, S, Wright, A.J, Noble, M.E.M, Mahoney, D.J, Campbell, I.D, Day, A.J, Jackson, D.G. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the Cd44-Hyaluronan Complex Provide Insight Into a Fundamental Carbohydrate-Protein Interaction.
Nat.Struct.Mol.Biol., 14, 2008
|
|
2JG2
 
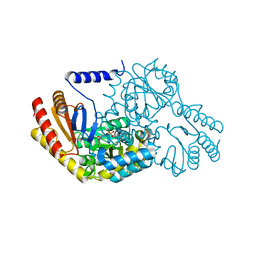 | | HIGH RESOLUTION STRUCTURE OF SPT WITH PLP INTERNAL ALDIMINE | | Descriptor: | MAGNESIUM ION, PYRIDOXAL-5'-PHOSPHATE, SERINE PALMITOYLTRANSFERASE | | Authors: | Yard, B.A, Carter, L.G, Johnson, K.A, Overton, I.M, Mcmahon, S.A, Dorward, M, Liu, H, Puech, D, Oke, M, Barton, G.J, Naismith, J.H, Campopiano, D.J. | | Deposit date: | 2007-02-07 | | Release date: | 2007-05-01 | | Last modified: | 2015-11-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Serine Palmitoyltransferase; Gateway to Sphingolipid Biosynthesis.
J.Mol.Biol., 370, 2007
|
|
2JYY
 
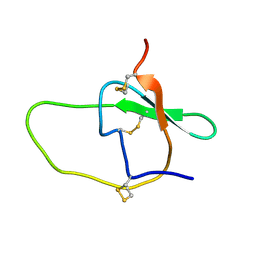 | |
2JSL
 
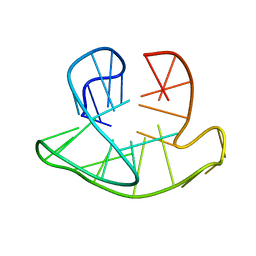 | | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, Form 2 Natural, NMR, 10 Structures | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-08 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution.
Nucleic Acids Res., 35, 2007
|
|
2JSQ
 
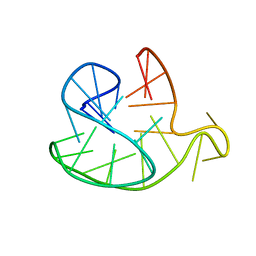 | | Monomeric Human Telomere DNA Tetraplex with 3+1 Strand Fold Topology, Two Edgewise Loops and Double-Chain Reversal Loop, Form 2 15BrG, NMR, 10 Structures | | Descriptor: | HUMAN TELOMERE DNA | | Authors: | Kuryavyi, V.V, Phan, A.T, Luu, K.N, Patel, D.J. | | Deposit date: | 2007-07-11 | | Release date: | 2008-07-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution.
Nucleic Acids Res., 35, 2007
|
|
2K2F
 
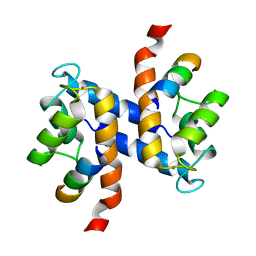 | |
1AV3
 
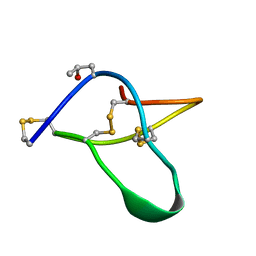 | | POTASSIUM CHANNEL BLOCKER KAPPA CONOTOXIN PVIIA FROM C. PURPURASCENS, NMR, 20 STRUCTURES | | Descriptor: | Kappa-conotoxin PVIIA | | Authors: | Scanlon, M.J, Naranjo, D, Thomas, L, Alewood, P.F, Lewis, R.J, Craik, D.J. | | Deposit date: | 1997-09-24 | | Release date: | 1998-10-14 | | Last modified: | 2020-12-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure and proposed binding mechanism of a novel potassium channel toxin kappa-conotoxin PVIIA.
Structure, 5, 1997
|
|
1B04
 
 | |
1B5K
 
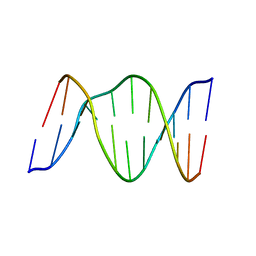 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE THYMIDINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*CP*EDCP*CP*AP*TP*GP*C)-3'), DNA (5'-D(*GP*CP*AP*TP*GP*TP*GP*TP*AP*CP*G)-3') | | Authors: | Cullinan, D, Korobka, A, Grollman, A.P, Patel, D.J, Eisenberg, M, De Santos, C.L. | | Deposit date: | 1999-01-07 | | Release date: | 1999-01-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite thymidine: comparison with the duplex containing deoxyadenosine opposite the adduct.
Biochemistry, 35, 1996
|
|
1AW4
 
 | |
1AX6
 
 | | SOLUTION STRUCTURE OF THE [AF]-C8-DG ADDUCT OPPOSITE A-2 DELETION SITE IN THE NARI HOT SPOT SEQUENCE CONTEXT; NMR, 6 STRUCTURES | | Descriptor: | 2-AMINOFLUORENE, DNA DUPLEX D(CTCGGC-[AF]G-CCATC)D(GATGGCCGAG) | | Authors: | Mao, B, Gorin, A.A, Gu, Z, Hingerty, B.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-30 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the aminofluorene-intercalated conformer of the syn [AF]-C8-dG adduct opposite a--2 deletion site in the NarI hot spot sequence context.
Biochemistry, 36, 1997
|
|
1AXV
 
 | | SOLUTION NMR STRUCTURE OF THE [BP]DA ADDUCT OPPOSITE DT IN A DNA DUPLEX, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(CTCTC-[BP]A-CTTCC)D(GGAAGTGAGAG) | | Authors: | Mao, B, Gu, Z, Gorin, A.A, Chen, J, Hingerty, B.E, Amid, S, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 1997-10-21 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the (+)-cis-anti-benzo[a]pyrene-dA ([BP]dA) adduct opposite dT in a DNA duplex.
Biochemistry, 38, 1999
|
|
1B6Y
 
 | | 3,N4-ETHENO-2'-DEOXYCYTIDINE OPPOSITE ADENINE IN AN 11-MER DUPLEX, SOLUTION STRUCTURE FROM NMR AND MOLECULAR DYNAMICS, 2 STRUCTURES | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*(EDC)P*CP*AP*TP*GP*C)-3', 5'-D(*GP*CP*AP*TP*GP*AP*GP*TP*AP*CP*G)-3' | | Authors: | Korobka, A, Cullinan, D, Cosman, M, Grollman, A.P, Patel, D.J, Eisenberg, M, De Los Santos, C. | | Deposit date: | 1999-01-19 | | Release date: | 1999-01-27 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an oligodeoxynucleotide duplex containing the exocyclic lesion 3,N4-etheno-2'-deoxycytidine opposite 2'-deoxyadenosine, determined by NMR spectroscopy and restrained molecular dynamics.
Biochemistry, 35, 1996
|
|
1BKF
 
 | | FK506 BINDING PROTEIN FKBP MUTANT R42K/H87V COMPLEX WITH IMMUNOSUPPRESSANT FK506 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Itoh, S, Decenzo, M.T, Livingston, D.J, Pearlman, D.A, Navia, M.A. | | Deposit date: | 1995-10-18 | | Release date: | 1996-08-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conformation of Fk506 in X-Ray Structures of its Complexes with Human Recombinant Fkbp12 Mutants
Bioorg.Med.Chem.Lett., 5, 1995
|
|
1BXX
 
 | |
1BIV
 
 | | BOVINE IMMUNODEFICIENCY VIRUS TAT-TAR COMPLEX, NMR, 5 STRUCTURES | | Descriptor: | TAR RNA, TAT PEPTIDE | | Authors: | Ye, X, Kumar, R.A, Patel, D.J. | | Deposit date: | 1996-06-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Molecular recognition in the bovine immunodeficiency virus Tat peptide-TAR RNA complex.
Chem.Biol., 2, 1995
|
|
