2CB4
 
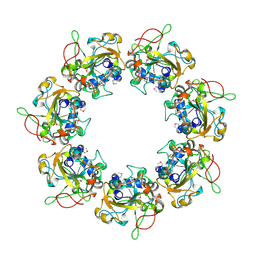 | | Crystal structure of the catalytic domain of the mosquitocidal toxin from Bacillus sphaericus, mutant E197Q | | Descriptor: | MOSQUITOCIDAL TOXIN | | Authors: | Reinert, D.J, Carpusca, I, Aktories, K, Schulz, G.E. | | Deposit date: | 2005-12-29 | | Release date: | 2006-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Mosquitocidal Toxin from Bacillus Sphaericus.
J.Mol.Biol., 357, 2006
|
|
1PT4
 
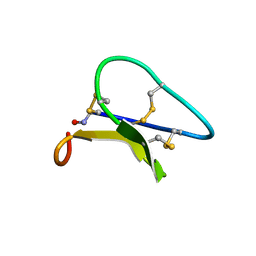 | | Solution structure of the Moebius cyclotide kalata B2 | | Descriptor: | kalata B2 | | Authors: | Jennings, C.V, Anderson, M.A, Daly, N.L, Rosengren, K.J, Craik, D.J. | | Deposit date: | 2003-06-23 | | Release date: | 2004-08-10 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Isolation, Solution Structure, and Insecticidal Activity of Kalata B2, a Circular Protein with a Twist: Do Mobius Strips Exist in Nature?(,)
Biochemistry, 44, 2005
|
|
6OI9
 
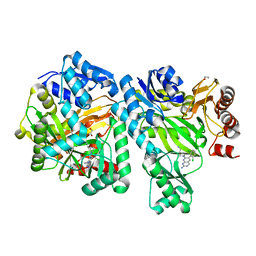 | | Crystal Structure of E. coli Biotin Carboxylase Complexed with 7-[3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine | | Descriptor: | 1,2-ETHANEDIOL, 7-[(3S)-3-(aminomethyl)pyrrolidin-1-yl]-6-(2,6-dichlorophenyl)pyrido[2,3-d]pyrimidin-2-amine, Biotin carboxylase | | Authors: | Andrews, L.D, Kane, T.R, Dozzo, P, Haglund, C.M, Hilderbrandt, D.J, Linsell, M.S, Machajewski, T, McEnroe, G, Serio, A.W, Wlasichuk, K.B, Neau, D.B, Pakhomova, S, Waldrop, G.L, Sharp, M, Pogliano, J, Cirz, R, Cohen, F. | | Deposit date: | 2019-04-09 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Optimization and Mechanistic Characterization of Pyridopyrimidine Inhibitors of Bacterial Biotin Carboxylase.
J.Med.Chem., 62, 2019
|
|
7OEU
 
 | | Model of open pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction | | Descriptor: | Haliangium ochraceum encapsulated ferritin, Linocin_M18 bacteriocin protein | | Authors: | Marles-Wright, J, Basle, A, Clarke, D.J, Ross, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Sci Adv, 8, 2022
|
|
1FYB
 
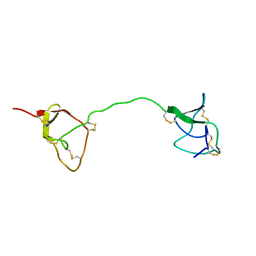 | | SOLUTION STRUCTURE OF C1-T1, A TWO-DOMAIN PROTEINASE INHIBITOR DERIVED FROM THE CIRCULAR PRECURSOR PROTEIN NA-PROPI FROM NICOTIANA ALATA | | Descriptor: | PROTEINASE INHIBITOR | | Authors: | Craik, D.J, Schirra, H.J, Scanlon, M.J, Anderson, M.A. | | Deposit date: | 2000-09-28 | | Release date: | 2001-02-21 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The solution structure of C1-T1, a two-domain proteinase inhibitor derived from a circular precursor protein from Nicotiana alata.
J.Mol.Biol., 306, 2001
|
|
6OJV
 
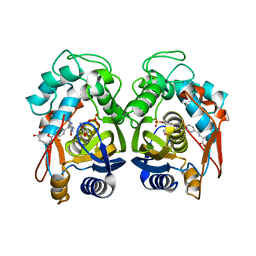 | | Crystal structure of human thymidylate synthase delta(7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-L-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-hydroxy-7H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzoyl}-L-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.593 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
6OJU
 
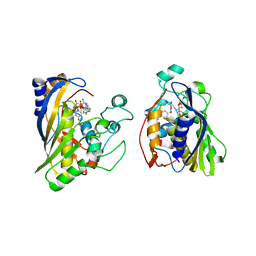 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}-D-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
7OE2
 
 | | Model of closed pentamer of the Haliangium ochraceum encapsulin from symmetry expansion of icosahedral single particle reconstruction | | Descriptor: | Haliangium ochraceum Encapsulated ferritin localisation sequence, Linocin_M18 bacteriocin protein | | Authors: | Marles-Wright, J, Basle, A, Clarke, D.J, Ross, J. | | Deposit date: | 2021-05-01 | | Release date: | 2022-02-09 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Pore dynamics and asymmetric cargo loading in an encapsulin nanocompartment.
Sci Adv, 8, 2022
|
|
1PFI
 
 | |
6OPF
 
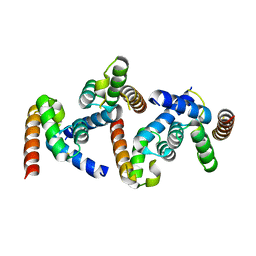 | |
1G6Q
 
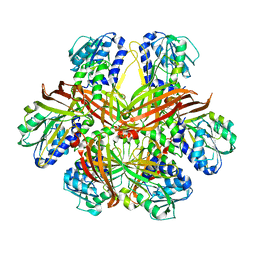 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | Descriptor: | HNRNP ARGININE N-METHYLTRANSFERASE | | Authors: | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
7ODW
 
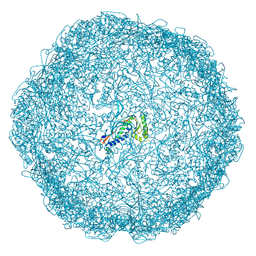 | |
1Q7R
 
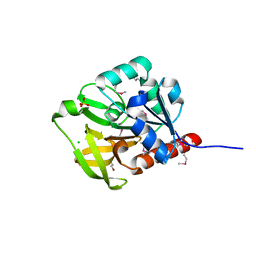 | |
1FVT
 
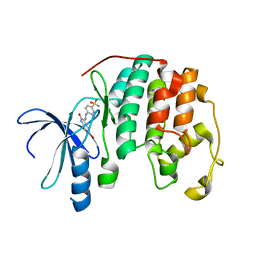 | | THE STRUCTURE OF CYCLIN-DEPENDENT KINASE 2 (CDK2) IN COMPLEX WITH AN OXINDOLE INHIBITOR | | Descriptor: | 4-[(2Z)-2-(5-bromo-2-oxo-1,2-dihydro-3H-indol-3-ylidene)hydrazinyl]benzene-1-sulfonamide, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Davis, S.T, Benson, B.G, Bramson, H.N, Chapman, D.E, Dickerson, S.H, Dold, K.M, Eberwein, D.J, Edelstein, M, Frye, S.V, Gampe Jr, R.T, Griffin, R.J, Harris, P.A, Hassell, A.M, Holmes, W.D, Hunter, R.N, Knick, V.B, Lackey, K, Lovejoy, B, Luzzio, M.J, Murray, D, Parker, P, Rocque, W.J, Shewchuk, L, Veal, J.M, Walker, D.H, Kuyper, L.K. | | Deposit date: | 2000-09-20 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Prevention of chemotherapy-induced alopecia in rats by CDK inhibitors.
Science, 291, 2001
|
|
1QXD
 
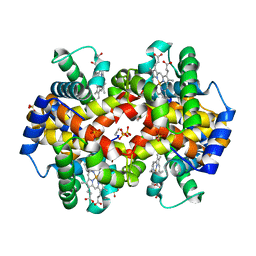 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
1G0Y
 
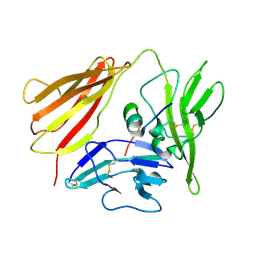 | | IL-1 RECEPTOR TYPE 1 COMPLEXED WITH ANTAGONIST PEPTIDE AF10847 | | Descriptor: | ANTAGONIST PEPTIDE AF10847, INTERLEUKIN-1 RECEPTOR, TYPE I | | Authors: | Vigers, G.P.A, Dripps, D.J, Edwards, C.K, Brandhuber, B.J. | | Deposit date: | 2000-10-09 | | Release date: | 2000-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of a small antagonist peptide bound to interleukin-1 receptor type 1.
J.Biol.Chem., 275, 2000
|
|
1R12
 
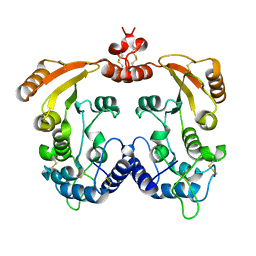 | | Native Aplysia ADP ribosyl cyclase | | Descriptor: | ADP-ribosyl cyclase | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-23 | | Release date: | 2004-03-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
1QXE
 
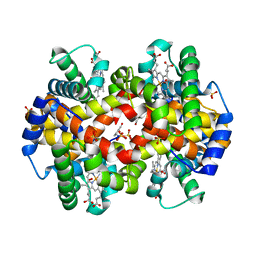 | | Structural Basis for the Potent Antisickling Effect of a Novel Class of 5-Membered Heterocyclic Aldehydic Compounds | | Descriptor: | 5-HYDROXYMETHYL-FURFURAL, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Safo, M.K, Abdulmalik, O, Danso-Danquah, R, Nokuri, S, Joshi, G.S, Musayev, F.N, Asakura, T, Abraham, D.J. | | Deposit date: | 2003-09-05 | | Release date: | 2003-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for the potent antisickling effect of a novel class of five-membered heterocyclic aldehydic compounds
J.Med.Chem., 47, 2004
|
|
6O6Y
 
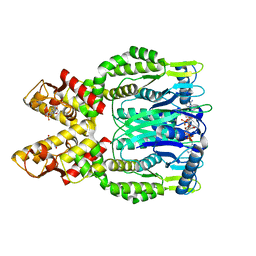 | | Crystal structure of Csm6 in complex with cyclic-tetraadenylates (cA4) by cocrystallization of Csm6 and cA4 | | Descriptor: | 2',3'- cyclic AMP, 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6O78
 
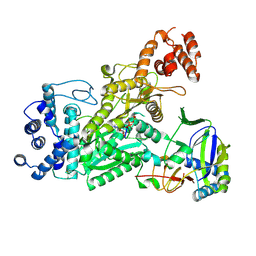 | | Crystal structure of Csm1-Csm4 cassette in complex with pppApApA | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, MANGANESE (II) ION, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
1G8X
 
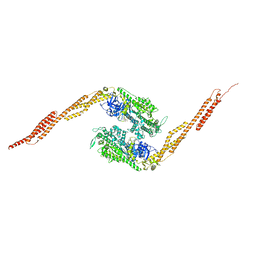 | | STRUCTURE OF A GENETICALLY ENGINEERED MOLECULAR MOTOR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, MYOSIN II HEAVY CHAIN FUSED TO ALPHA-ACTININ 3 | | Authors: | Kliche, W, Fujita-Becker, S, Kollmar, M, Manstein, D.J, Kull, F.J. | | Deposit date: | 2000-11-21 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a genetically engineered molecular motor.
EMBO J., 20, 2001
|
|
6ONJ
 
 | |
1R0S
 
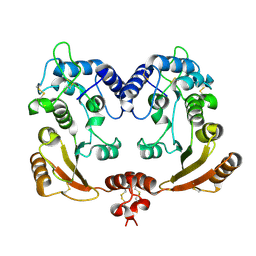 | | Crystal structure of ADP-ribosyl cyclase Glu179Ala mutant | | Descriptor: | ADP-ribosyl cyclase | | Authors: | Love, M.L, Szebenyi, D.M.E, Kriksunov, I.A, Thiel, D.J, Munshi, C, Graeff, R, Lee, H.C, Hao, Q. | | Deposit date: | 2003-09-22 | | Release date: | 2004-03-09 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | ADP-ribosyl cyclase; crystal structures reveal a covalent intermediate.
Structure, 12, 2004
|
|
7OG1
 
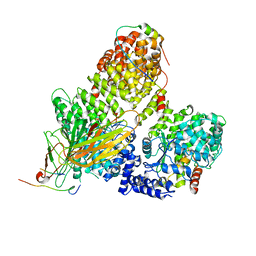 | | AP2 clathrin adaptor core in complex with cargo peptide and FCHO2 | | Descriptor: | AP-2 complex subunit alpha-2, AP-2 complex subunit beta, AP-2 complex subunit mu, ... | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
7OFP
 
 | | Apo Structure of Mu2 Adaptin Subunit (Ap50) Of AP2 Clathrin Adaptor | | Descriptor: | AP-2 complex subunit mu, CITRIC ACID, GLYCEROL | | Authors: | Zaccai, N.R, Kelly, B.T, Evans, P.R, Owen, D.J. | | Deposit date: | 2021-05-05 | | Release date: | 2022-06-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | FCHO controls AP2's initiating role in endocytosis through a PtdIns(4,5)P 2 -dependent switch.
Sci Adv, 8, 2022
|
|
